Related Research Articles

Sea urchins are spiny, globular echinoderms in the class Echinoidea. About 950 species of sea urchin are distributed on the seabeds of every ocean and inhabit every depth zone from the intertidal seashore down to 5,000 meters. The spherical, hard shells (tests) of sea urchins are round and covered in spines. Most urchin spines range in length from 3 to 10 cm, with outliers such as the black sea urchin possessing spines as long as 30 cm (12 in). Sea urchins move slowly, crawling with tube feet, and also propel themselves with their spines. Although algae are the primary diet, sea urchins also eat slow-moving (sessile) animals. Predators that eat sea urchins include a wide variety of fish, starfish, crabs, marine mammals, and humans.
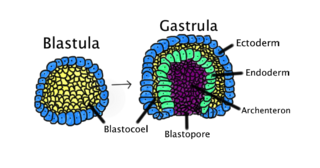
Gastrulation is the stage in the early embryonic development of most animals, during which the blastula, or in mammals the blastocyst is reorganized into a two-layered or three-layered embryo known as the gastrula. Before gastrulation, the embryo is a continuous epithelial sheet of cells; by the end of gastrulation, the embryo has begun differentiation to establish distinct cell lineages, set up the basic axes of the body, and internalized one or more cell types including the prospective gut.

The blastocoel, also spelled blastocoele and blastocele, and also called cleavage cavity, or segmentation cavity is a fluid-filled or yolk-filled cavity that forms in the blastula during very early embryonic development. At this stage in mammals the blastula develops into the blastocyst containing an inner cell mass, and outer trophectoderm.

Sir Richard Timothy Hunt, is a British biochemist and molecular physiologist. He was awarded the 2001 Nobel Prize in Physiology or Medicine with Paul Nurse and Leland H. Hartwell for their discoveries of protein molecules that control the division of cells. While studying fertilized sea urchin eggs in the early 1980s, Hunt discovered cyclin, a protein that cyclically aggregates and is depleted during cell division cycles.

Proteinase 3, also known as PRTN3, is an enzyme that in humans is encoded by the PRTN3 gene.
Aspergillopepsin I is an enzyme. This enzyme catalyses the following chemical reaction
Clostripain is a proteinase that cleaves proteins on the carboxyl peptide bond of arginine. It was isolated from Clostridium histolyticum. The isoelectric point of the enzyme is 4.8-4.9, and optimum pH is 7.4~7.8. The composition of the enzyme is indicated to be of two chains of relative molecular mass 45,000 and 12,500.
In biochemistry, sulfatases EC 3.1.6.- are a class of enzymes of the esterase class that catalyze the hydrolysis of sulfate esters into an alcohol and a bisulfate:

Cathepsin G is a protein that in humans is encoded by the CTSG gene. It is one of the three serine proteases of the chymotrypsin family that are stored in the azurophil granules, and also a member of the peptidase S1 protein family. Cathepsin G plays an important role in eliminating intracellular pathogens and breaking down tissues at inflammatory sites, as well as in anti-inflammatory response.

Thermolysin is a thermostable neutral metalloproteinase enzyme produced by the Gram-positive bacteria Bacillus thermoproteolyticus. It requires one zinc ion for enzyme activity and four calcium ions for structural stability. Thermolysin specifically catalyzes the hydrolysis of peptide bonds containing hydrophobic amino acids. However thermolysin is also widely used for peptide bond formation through the reverse reaction of hydrolysis. Thermolysin is the most stable member of a family of metalloproteinases produced by various Bacillus species. These enzymes are also termed 'neutral' proteinases or thermolysin -like proteinases (TLPs).
Susan G. Ernst is professor emerita at Tufts University known for her work on cell development using sea urchins as a model system. She is an elected fellow of the American Association for the Advancement of Science.

Cathepsin E is an enzyme that in humans is encoded by the CTSE gene. The enzyme is also known as slow-moving proteinase, erythrocyte membrane aspartic proteinase, SMP, EMAP, non-pepsin proteinase, cathepsin D-like acid proteinase, cathepsin E-like acid proteinase, cathepsin D-type proteinase) is an enzyme.
In evolutionary developmental biology, inversion refers to the hypothesis that during the course of animal evolution, the structures along the dorsoventral (DV) axis have taken on an orientation opposite that of the ancestral form.
Lysyl endopeptidase is an enzyme. This enzyme catalyses the following chemical reaction
Glycyl endopeptidase is an enzyme. This enzyme catalyses the following chemical reaction
Serralysin is an enzyme. This enzyme catalyses the following chemical reaction
Bacillolysin is an enzyme. This enzyme catalyses the following chemical reaction
Choriolysin H is an enzyme. This enzyme catalyses the following chemical reaction
Donna R. Maglott is a staff scientist at the National Center for Biotechnology Information known for her research on large-scale genomics projects, including the mouse genome and development of databases required for genomics research.
Cytochrome P450, family 27, also known as CYP27, is a Deuterostome cytochrome P450 monooxygenase family found in human genome. This family belongs to Mitochondrial clan CYPs, which is located in the inner membrane of mitochondria(IMM). There are three members in the human genome, CYP27A1, CYP27B1 and CYP27C1, and an ortholog CYP27F1 in sea urchins, Strongylocentrotus purpuratus.
References
- ↑ Barrett D, Edwards BF (1976). Hatching enzyme of the sea urchin Strongylocentrotus purpuratus. Methods in Enzymology. Vol. 45. pp. 354–73. doi:10.1016/s0076-6879(76)45032-2. PMID 1012003.
- ↑ Lepage T, Gache C (March 1989). "Purification and characterization of the sea urchin embryo hatching enzyme". The Journal of Biological Chemistry. 264 (9): 4787–93. doi: 10.1016/S0021-9258(18)83659-6 . PMID 2925668.
- ↑ Lepage T, Gache C (September 1990). "Early expression of a collagenase-like hatching enzyme gene in the sea urchin embryo". The EMBO Journal. 9 (9): 3003–12. doi:10.1002/j.1460-2075.1990.tb07493.x. PMC 552018 . PMID 2167841.
- ↑ Nomura K, Tanaka H, Kikkawa Y, Yamaguchi M, Suzuki N (June 1991). "The specificity of sea urchin hatching enzyme (envelysin) places it in the mammalian matrix metalloproteinase family". Biochemistry. 30 (25): 6115–23. doi:10.1021/bi00239a005. PMID 1711895.