| MHC class I, A | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (heterodimer) | ||||||||||
 Illustration of HLA-A | ||||||||||
| Protein type | Transmembrane protein | |||||||||
| Function | Peptide presentation for immune recognition | |||||||||
| ||||||||||
HLA-A is a group of human leukocyte antigens (HLA) that are encoded by the HLA-A locus, which is located at human chromosome 6p21.3. [1] HLA is a major histocompatibility complex (MHC) antigen specific to humans. HLA-A is one of three major types of human MHC class I transmembrane proteins. The others are HLA-B and HLA-C. [2] The protein is a heterodimer, and is composed of a heavy α chain and smaller β chain. The α chain is encoded by a variant HLA-A gene, and the β chain (β2-microglobulin) is an invariant β2 microglobulin molecule. [3] The β2 microglobulin protein is encoded by the B2M gene, [4] which is located at chromosome 15q21.1 in humans. [5]
Contents
- HLA-A gene
- Alleles
- Protein
- Function
- Natural function
- Other activities
- Role in disease
- HIV/AIDS
- Summary
- References
- External links
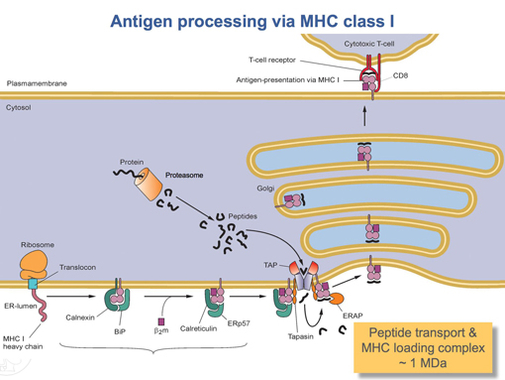
MHC Class I molecules such as HLA-A participate in a process that presents short polypeptides to the immune system. These polypeptides are typically 7–11 amino acids in length and originate from proteins being expressed by the cell. There are two classes of polypeptide that can be presented by an HLA protein: those that are supposed to be expressed by the cell (self) and those of foreign derivation (non-self). [6] Under normal conditions cytotoxic T cells, which normally patrol the body in the blood, "read" the peptide presented by the complex. T cells, if functioning properly, only bind to non-self peptides. If binding occurs, a series of events is initiated culminating in cell death via apoptosis. [7] In this manner, the human body eliminates any cells infected by a virus or expressing proteins they shouldn't be (e.g. cancerous cells).
For humans, as in most mammalian populations, MHC Class I molecules are extremely variable in their primary structure, and HLA-A is ranked among the genes with the fastest-evolving coding sequence in humans. As of March 2022, there are 7,452 known HLA-A alleles coding for 4,305 active proteins and 375 null proteins.This level of variation on MHC Class I is the primary cause of transplant rejection, as random transplantation between donor and host is unlikely to result in a matching of HLA-A, B or C antigens. Evolutionary biologists also believe that the wide variation in HLAs is a result of a balancing act between conflicting pathogenic pressures. Greater variety of HLAs decreases the probability that the entire population will be wiped out by a single pathogen as certain individuals will be highly resistant to each pathogen. [6] The effect of HLA-A variation on HIV/AIDS progression is discussed below.