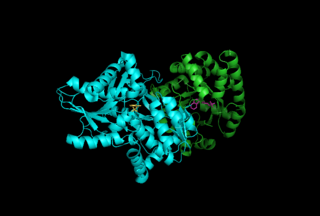
Tryptophan synthase or tryptophan synthetase is an enzyme that catalyzes the final two steps in the biosynthesis of tryptophan. It is commonly found in Eubacteria, Archaebacteria, Protista, Fungi, and Plantae. However, it is absent from Animalia. It is typically found as an α2β2 tetramer. The α subunits catalyze the reversible formation of indole and glyceraldehyde-3-phosphate (G3P) from indole-3-glycerol phosphate (IGP). The β subunits catalyze the irreversible condensation of indole and serine to form tryptophan in a pyridoxal phosphate (PLP) dependent reaction. Each α active site is connected to a β active site by a 25 Ångstrom long hydrophobic channel contained within the enzyme. This facilitates the diffusion of indole formed at α active sites directly to β active sites in a process known as substrate channeling. The active sites of tryptophan synthase are allosterically coupled.
Shikimic acid, more commonly known as its anionic form shikimate, is a cyclohexene, a cyclitol and a cyclohexanecarboxylic acid. It is an important biochemical metabolite in plants and microorganisms. Its name comes from the Japanese flower shikimi, from which it was first isolated in 1885 by Johan Fredrik Eykman. The elucidation of its structure was made nearly 50 years later.
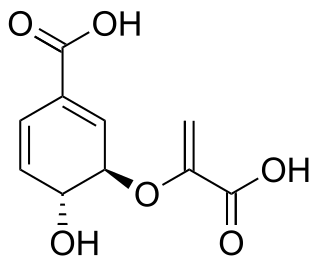
Chorismic acid, more commonly known as its anionic form chorismate, is an important biochemical intermediate in plants and microorganisms. It is a precursor for:

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.

Amino acid biosynthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids).

Carbamoyl phosphate synthetase catalyzes the ATP-dependent synthesis of carbamoyl phosphate from glutamine or ammonia and bicarbonate. This enzyme catalyzes the reaction of ATP and bicarbonate to produce carboxy phosphate and ADP. Carboxy phosphate reacts with ammonia to give carbamic acid. In turn, carbamic acid reacts with a second ATP to give carbamoyl phosphate plus ADP.

Isochorismate synthase ( EC 5.4.4.2) is an isomerase enzyme that catalyzes the first step in the biosynthesis of vitamin K2 (menaquinone) in Escherichia coli.

In enzymology, a phosphoribosylanthranilate isomerase (PRAI) is an enzyme that catalyzes the third step of the synthesis of the amino acid tryptophan.

The enzyme anthranilate synthase catalyzes the chemical reaction
The enzyme indole-3-glycerol-phosphate lyase catalyzes the chemical reaction
The enzyme indolepyruvate decarboxylase (EC 4.1.1.74) catalyzes the chemical reaction
The enzyme L-fuculose-phosphate aldolase (EC 4.1.2.17) catalyzes the chemical reaction

In enzymology, an aminodeoxychorismate synthase is an enzyme that catalyzes the chemical reaction

The enzyme 3-dehydroquinate synthase catalyzes the chemical reaction
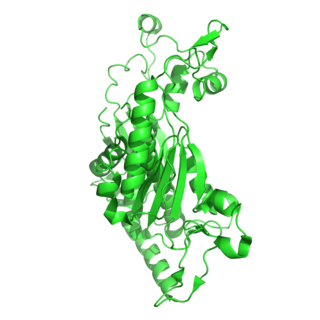
The enzyme chorismate synthase catalyzes the chemical reaction
In enzymology, a 3-deoxy-8-phosphooctulonate synthase (EC 2.5.1.55) is an enzyme that catalyzes the chemical reaction

The FPG IleRS zinc finger domain represents a zinc finger domain found at the C-terminal in both DNA glycosylase/AP lyase enzymes and in isoleucyl tRNA synthetase. In these two types of enzymes, the C-terminal domain forms a zinc finger.
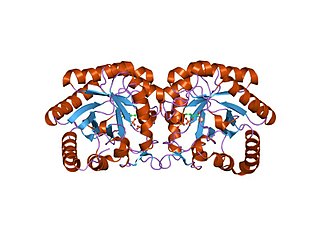
3-Deoxy-D-arabinoheptulosonate 7-phosphate (DAHP) synthase is the first enzyme in a series of metabolic reactions known as the shikimate pathway, which is responsible for the biosynthesis of the amino acids phenylalanine, tyrosine, and tryptophan. Since it is the first enzyme in the shikimate pathway, it controls the amount of carbon entering the pathway. Enzyme inhibition is the primary method of regulating the amount of carbon entering the pathway. Forms of this enzyme differ between organisms, but can be considered DAHP synthase based upon the reaction that is catalyzed by this enzyme.

3-Deoxy-D-arabino-heptulosonic acid 7-phosphate (DAHP) is a 7-carbon ulosonic acid. This compound is found in the shikimic acid biosynthesis pathway and is an intermediate in the production of aromatic amino acids.
3-hexulose-6-phosphate synthase is an enzyme with systematic name D-arabino-hex-3-ulose-6-phosphate formaldehyde-lyase (D-ribulose-5-phosphate-forming). This enzyme catalyses the following chemical reaction
















