Related Research Articles
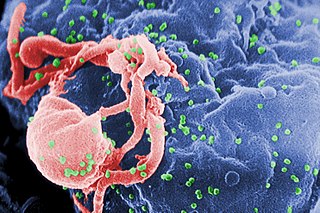
The human immunodeficiency viruses (HIV) are two species of Lentivirus that infect humans. Over time, they cause acquired immunodeficiency syndrome (AIDS), a condition in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive. Without treatment, average survival time after infection with HIV is estimated to be 9 to 11 years, depending on the HIV subtype. In most cases, HIV is a sexually transmitted infection and occurs by contact with or transfer of blood, pre-ejaculate, semen, and vaginal fluids. Research has shown that HIV is untransmittable through condomless sexual intercourse if the HIV-positive partner has a consistently undetectable viral load. Non-sexual transmission can occur from an infected mother to her infant during pregnancy, during childbirth by exposure to her blood or vaginal fluid, and through breast milk. Within these bodily fluids, HIV is present as both free virus particles and virus within infected immune cells.

Simian immunodeficiency virus (SIV) is a species of retrovirus that cause persistent infections in at least 45 species of African non-human primates. Based on analysis of strains found in four species of monkeys from Bioko Island, which was isolated from the mainland by rising sea levels about 11,000 years ago, it has been concluded that SIV has been present in monkeys and apes for at least 32,000 years, and probably much longer.

Tripartite motif-containing protein 5 also known as RING finger protein 88 is a protein that in humans is encoded by the TRIM5 gene. The alpha isoform of this protein, TRIM5α, is a retrovirus restriction factor, which mediates species-specific, early block to retrovirus infection.
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species. The genus includes the human immunodeficiency virus (HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals.
The genome and proteins of HIV have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

Viral infectivity factor, or Vif, is an accessory protein found in HIV and other lentiviruses. Its role is to disrupt the antiviral activity of the human enzyme APOBEC by targeting it for ubiquitination and cellular degradation. APOBEC is a cytidine deaminase enzyme that mutates viral nucleic acids.
Human foamy virus (HFV) is a retrovirus and specifically belongs to the genus Spumavirus. The spumaviruses are complex and significantly different from the other six genera of retroviruses in several ways. The foamy viruses derive their name from the characteristic ‘foamy’ appearance of the cytopathic effect (CPE) induced in the cells. Foamy virus in humans occurs only as a result of zoonotic infection.

APOBEC3G is a human enzyme encoded by the APOBEC3G gene that belongs to the APOBEC superfamily of proteins. This family of proteins has been suggested to play an important role in innate anti-viral immunity. APOBEC3G belongs to the family of cytidine deaminases that catalyze the deamination of cytidine to uridine in the single stranded DNA substrate. The C-terminal domain of A3G renders catalytic activity, several NMR and crystal structures explain the substrate specificity and catalytic activity.
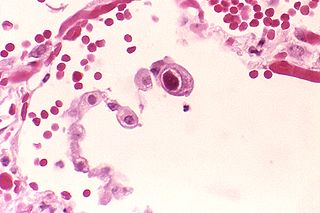
Human betaherpesvirus 5, also called human cytomegalovirus (HCMV), is species of virus in the genus Cytomegalovirus, which in turn is a member of the viral family known as Herpesviridae or herpesviruses. It is also commonly called CMV. Within Herpesviridae, HCMV belongs to the Betaherpesvirinae subfamily, which also includes cytomegaloviruses from other mammals. CMV is a double-stranded DNA virus.

Proteasome activator complex subunit 4 is a protein that in humans is encoded by the PSME4 gene.

Mitochondrial antiviral-signaling protein (MAVS) is a protein that is essential for antiviral innate immunity. MAVS is located in the inner membrane of the mitochondria, peroxisomes, and endoplasmic reticulum (ER). Upon viral infection, a group of cytosolic proteins will detect the presence of the virus and bind to MAVS, thereby activating MAVS. The activation of MAVS leads the virally infected cell to secrete cytokines. This induces an immune response which kills the host's virally infected cells, resulting in clearance of the virus.

DNA dC->dU-editing enzyme APOBEC-3C is a protein that in humans is encoded by the APOBEC3C gene.

Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A, also known as APOBEC3A, or A3A is a gene of the APOBEC3 family found in humans, non-human primates, and some other mammals. It is a single-domain DNA cytidine deaminase with antiviral effects. While other members of the family such as APOBEC3G are believed to act by editing ssDNA by removing an amino group from cytosine in DNA, introducing a cytosine to uracil change which can ultimately lead to a cytosine to thymine mutation, one study suggests that APOBEC3A can inhibit parvoviruses by another mechanism. The cellular function of APOBEC3A is likely to be the destruction of foreign DNA through extensive deamination of cytosine.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. "APOBEC3 proteins mediate the clearance of foreign DNA from human cells". Nature Structural & Molecular Biology. 17 (2): 222–9. doi:10.1038/nsmb.1744. PMC 2921484. PMID 20062055.

Tetherin, also known as bone marrow stromal antigen 2, is a lipid raft associated protein that in humans is encoded by the BST2 gene. In addition, tetherin has been designated as CD317. This protein is constitutively expressed in mature B cells, plasma cells and plasmacytoid dendritic cells, and in many other cells, it is only expressed as a response to stimuli from IFN pathway.

DNA dC->dU-editing enzyme APOBEC-3H, also known as Apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3H or APOBEC-related protein 10, is a protein that in humans is encoded by the APOBEC3H gene.

Virus inhibitory protein, endoplasmic reticulum-associated, interferon-inducible (Viperin), also known as RSAD2, is a protein that is encoded by the RSAD2 gene. Viperin is a multifunctional protein in viral processes that is an interferon stimulated gene. It has been reported that viperin could be induced by either IFN-dependent or IFN-independent pathways and certain viruses may use viperin to increase their infectivity.

Stimulator of interferon genes (STING), also known as transmembrane protein 173 (TMEM173) and MPYS/MITA/ERIS is a protein that in humans is encoded by the STING1 gene.
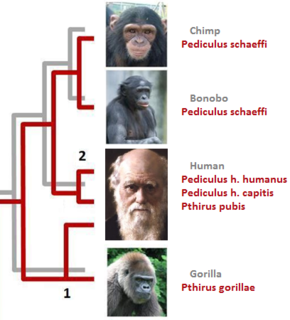
In parasitology and epidemiology, a host switch is an evolutionary change of the host specificity of a parasite or pathogen. For example, the human immunodeficiency virus used to infect and circulate in non-human primates in West-central Africa, but switched to humans in the early 20th century.
HSV epigenetics is the epigenetic modification of herpes simplex virus (HSV) genetic code.
Theodora Hatziioannou is a Greek-American virologist. She known for her work discovering restriction factors that counteract HIV-AIDS and other primate lentiviruses, thus restricting them to specific species, and making it hard to study HIV-1 in animals. Her findings allowed her to develop the first HIV-1-based virus capable of recapitulating AIDS-like symptoms in a non-hominid. She is a Research Associate Professor in the Laboratory of Retrovirology at The Rockefeller University in New York. She is a co-author of a textbook on virology, Principles of Virology.
References
- ↑ Yan N, Chen ZJ (February 2012). "Intrinsic antiviral immunity". Nature Immunology. 13 (3): 214–22. doi:10.1038/ni.2229. PMC 3549670 . PMID 22344284.
- ↑ Bieniasz PD (November 2004). "Intrinsic immunity: a front-line defense against viral attack". Nat. Immunol. 5 (11): 1109–15. doi:10.1038/ni1125. PMID 15496950. S2CID 8671718.
- ↑ Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J (February 2004). "The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys". Nature. 427 (6977): 848–53. Bibcode:2004Natur.427..848S. doi:10.1038/nature02343. PMID 14985764. S2CID 1801120.
- ↑ Sheehy AM, Gaddis NC, Choi JD, Malim MH (August 2002). "Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein". Nature. 418 (6898): 646–50. Bibcode:2002Natur.418..646S. doi:10.1038/nature00939. PMID 12167863. S2CID 4403228.
- ↑ Saffert RT, Kalejta RF (April 2006). "Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression". J. Virol. 80 (8): 3863–71. doi:10.1128/JVI.80.8.3863-3871.2006. PMC 1440479 . PMID 16571803.