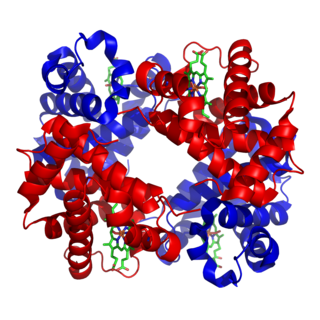
Metalloprotein is a generic term for a protein that contains a metal ion cofactor. A large proportion of all proteins are part of this category. For instance, at least 1000 human proteins contain zinc-binding protein domains although there may be up to 3000 human zinc metalloproteins.

In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the binding site, and residues that catalyse a reaction of that substrate, the catalytic site. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes.

Metallothionein (MT) is a family of cysteine-rich, low molecular weight proteins. They are localized to the membrane of the Golgi apparatus. MTs have the capacity to bind both physiological and xenobiotic heavy metals through the thiol group of its cysteine residues, which represent nearly 30% of its constituent amino acid residues.
Thioredoxin reductases are enzymes that reduce thioredoxin (Trx). Two classes of thioredoxin reductase have been identified: one class in bacteria and some eukaryotes and one in animals. In bacteria TrxR also catalyzes the reduction of glutaredoxin like proteins known as NrdH. Both classes are flavoproteins which function as homodimers. Each monomer contains a FAD prosthetic group, a NADPH binding domain, and an active site containing a redox-active disulfide bond.
Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Flavoproteins are proteins that contain a nucleic acid derivative of riboflavin. These proteins are involved in a wide array of biological processes, including removal of radicals contributing to oxidative stress, photosynthesis, and DNA repair. The flavoproteins are some of the most-studied families of enzymes.
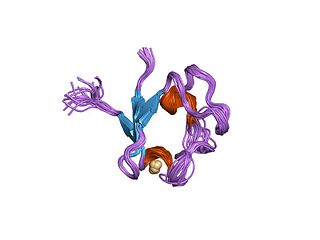
Rubredoxins are a class of low-molecular-weight iron-containing proteins found in sulfur-metabolizing bacteria and archaea. Sometimes rubredoxins are classified as iron-sulfur proteins; however, in contrast to iron-sulfur proteins, rubredoxins do not contain inorganic sulfide. Like cytochromes, ferredoxins and Rieske proteins, rubredoxins are thought to participate in electron transfer in biological systems. Recent work in bacteria and algae have led to the hypothesis that some rubredoxins may instead have a role in delivering iron to metalloproteins.
Copper proteins are proteins that contain one or more copper ions as prosthetic groups. Copper proteins are found in all forms of air-breathing life. These proteins are usually associated with electron-transfer with or without the involvement of oxygen (O2). Some organisms even use copper proteins to carry oxygen instead of iron proteins. A prominent copper proteins in humans is in cytochrome c oxidase (cco). The enzyme cco mediates the controlled combustion that produces ATP.
Nitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2− to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.
Oxidative protein folding is a process that is responsible for the formation of disulfide bonds between cysteine residues in proteins. The driving force behind this process is a redox reaction, in which electrons pass between several proteins and finally to a terminal electron acceptor.

Nitrate reductases are molybdoenzymes that reduce nitrate to nitrite. This reaction is critical for the production of protein in most crop plants, as nitrate is the predominant source of nitrogen in fertilized soils.
Nitrile hydratases are mononuclear iron or non-corrinoid cobalt enzymes that catalyse the hydration of diverse nitriles to their corresponding amides

Matrilysin also known as matrix metalloproteinase-7 (MMP-7), pump-1 protease (PUMP-1), or uterine metalloproteinase is an enzyme in humans that is encoded by the MMP7 gene. The enzyme has also been known as matrin, putative metalloproteinase-1, matrix metalloproteinase pump 1, PUMP-1 proteinase, PUMP, metalloproteinase pump-1, putative metalloproteinase, MMP). Human MMP-7 has a molecular weight around 30 kDa.
In enzymology, a ferredoxin-NADP+ reductase (EC 1.18.1.2) abbreviated FNR, is an enzyme that catalyzes the chemical reaction

Thiosulfate dehydrogenase is an enzyme that catalyzes the chemical reaction:

In molecular biology, the ars operon is an operon found in several bacterial taxon. It is required for the detoxification of arsenate, arsenite, and antimonite. This system transports arsenite and antimonite out of the cell. The pump is composed of two polypeptides, the products of the arsA and arsB genes. This two-subunit enzyme produces resistance to arsenite and antimonite. Arsenate, however, must first be reduced to arsenite before it is extruded. A third gene, arsC, expands the substrate specificity to allow for arsenate pumping and resistance. ArsC is an approximately 150-residue arsenate reductase that uses reduced glutathione (GSH) to convert arsenate to arsenite with a redox active cysteine residue in the active site. ArsC forms an active quaternary complex with GSH, arsenate, and glutaredoxin 1 (Grx1). The three ligands must be present simultaneously for reduction to occur.
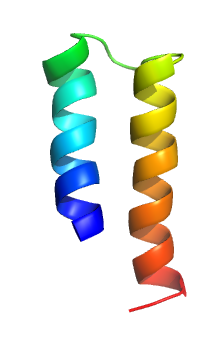
The mercury transporter superfamily is a family of transmembrane bacterial transporters of mercury ions. The common origin of all Mer superfamily members has been established. The common elements between family members are included in TMSs 1-2. A representative list of the subfamilies and proteins that belong to those subfamilies is available in the Transporter Classification Database.

Nickel superoxide dismutase (Ni-SOD) is a metalloenzyme that, like the other superoxide dismutases, protects cells from oxidative damage by catalyzing the disproportionation of the cytotoxic superoxide radical to hydrogen peroxide and molecular oxygen. Superoxide is a reactive oxygen species that is produced in large amounts during photosynthesis and aerobic cellular respiration. The equation for the disproportionation of superoxide is shown below:

L-ornithine N5 monooxygenase (EC 1.14.13.195 or EC 1.14.13.196) is an enzyme which catalyzes one of the following chemical reactions:
L-ornithine + NADPH + O2 N(5)-hydroxy-L-ornithine + NADP+ + H2O L-ornithine + NAD(P)H + O2 N(5)-hydroxy-L-ornithine + NAD(P)+ + H2O














