Related Research Articles

Ornithine transcarbamylase (OTC) is an enzyme that catalyzes the reaction between carbamoyl phosphate (CP) and ornithine (Orn) to form citrulline (Cit) and phosphate (Pi). There are two classes of OTC: anabolic and catabolic. This article focuses on anabolic OTC. Anabolic OTC facilitates the sixth step in the biosynthesis of the amino acid arginine in prokaryotes. In contrast, mammalian OTC plays an essential role in the urea cycle, the purpose of which is to capture toxic ammonia and transform it into urea, a less toxic nitrogen source, for excretion.
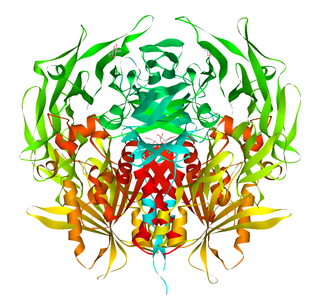
Dipeptidyl peptidase-4 (DPP4), also known as adenosine deaminase complexing protein 2 or CD26 is a protein that, in humans, is encoded by the DPP4 gene. DPP4 is related to FAP, DPP8, and DPP9. The enzyme was discovered in 1966 by Hopsu-Havu and Glenner, and as a result of various studies on chemism, was called dipeptidyl peptidase IV [DP IV].

Methylmalonyl-CoA mutase (EC 5.4.99.2, MCM), mitochondrial, also known as methylmalonyl-CoA isomerase, is a protein that in humans is encoded by the MUT gene. This vitamin B12-dependent enzyme catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA in humans. Mutations in MUT gene may lead to various types of methylmalonic aciduria.
In molecular biology, the Signal Peptide Peptidase (SPP) is a type of protein that specifically cleaves parts of other proteins. It is an intramembrane aspartyl protease with the conserved active site motifs 'YD' and 'GxGD' in adjacent transmembrane domains (TMDs). Its sequences is highly conserved in different vertebrate species. SPP cleaves remnant signal peptides left behind in membrane by the action of signal peptidase and also plays key roles in immune surveillance and the maturation of certain viral proteins.

Propionyl-CoA carboxylase (EC 6.4.1.3, PCC) catalyses the carboxylation reaction of propionyl-CoA in the mitochondrial matrix. PCC has been classified both as a ligase and a lyase. The enzyme is biotin-dependent. The product of the reaction is (S)-methylmalonyl CoA.
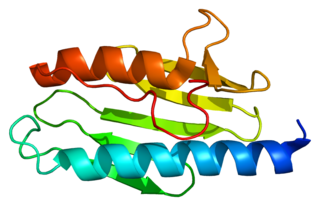
Frataxin is a protein that in humans is encoded by the FXN gene.

Met-enkephalin, also known as metenkefalin (INN), sometimes referred to as opioid growth factor (OGF), is a naturally occurring, endogenous opioid peptide that has opioid effects of a relatively short duration. It is one of the two forms of enkephalin, the other being leu-enkephalin. The enkephalins are considered to be the primary endogenous ligands of the δ-opioid receptor, due to their high potency and selectivity for the site over the other endogenous opioids.
Kexin is a prohormone-processing protease, specifically a yeast serine peptidase, found in the budding yeast. It catalyzes the cleavage of -Lys-Arg- and -Arg-Arg- bonds to process yeast alpha-factor pheromone and killer toxin precursors. The human homolog is PCSK4. It is a family of subtilisin-like peptidases. Even though there are a few prokaryote kexin-like peptidases, all kexins are eukaryotes. The enzyme is encoded by the yeast gene KEX2, and usually referred to in the scientific community as Kex2p. It shares structural similarities with the bacterial protease subtilisin. The first mammalian homologue of this protein to be identified was furin. In the mammal, kexin-like peptidases function in creating and regulating many differing proproteins.

Tripeptidyl-peptidase 2 is an enzyme that in humans is encoded by the TPP2 gene. Among other things it is heavily implicated in MHC (HLA) class-I processing, as it has both endopeptidase and exopeptidase activity.

Pitrilysin metallopeptidase 1 also known as presequence protease, mitochondrial (PreP) and metalloprotease 1 (MTP-1) is an enzyme that in humans is encoded by the PITRM1 gene. It is also sometimes called metalloprotease 1 (MP1).PreP facilitates proteostasis by utilizing an ~13300-A(3) catalytic chamber to degrade toxic peptides, including mitochondrial presequences and β-amyloid. Deficiency of PreP is found associated with Alzheimer’s disease. Reduced levels of PreP via RNAi mediated knockdown have been shown to lead to defective maturation of the protein Frataxin.

NAD-dependent deacetylase sirtuin-3, mitochondrial also known as SIRT3 is a protein that in humans is encoded by the SIRT3 gene [sirtuin 3 ]. SIRT3 is member of the mammalian sirtuin family of proteins, which are homologs to the yeast Sir2 protein. SIRT3 exhibits NAD+-dependent deacetylase activity.

Mitochondrial-processing peptidase subunit beta is an enzyme that in humans is encoded by the PMPCB gene. This gene is a member of the peptidase M16 family and encodes a protein with a zinc-binding motif. This protein is located in the mitochondrial matrix and catalyzes the cleavage of the leader peptides of precursor proteins newly imported into the mitochondria, though it only functions as part of a heterodimeric complex.

Mitochondrial-processing peptidase subunit alpha is an enzyme that in humans is encoded by the PMPCA gene. This gene PMPCA encoded a protein that is a member of the peptidase M16 family. This protein is located in the mitochondrial matrix and catalyzes the cleavage of the leader peptides of precursor proteins newly imported into the mitochondria, though it only functions as part of a heterodimeric complex.

Alpha-aminoadipic semialdehyde synthase is an enzyme encoded by the AASS gene in humans and is involved in their major lysine degradation pathway. It is similar to the separate enzymes coded for by the LYS1 and LYS9 genes in yeast, and related to, although not similar in structure, the bifunctional enzyme found in plants. In humans, mutations in the AASS gene, and the corresponding alpha-aminoadipic semialdehyde synthase enzyme are associated with familial hyperlysinemia. This condition is inherited in an autosomal recessive pattern and is not considered a particularly negative condition, thus making it a rare disease.

ATP-dependent Clp protease proteolytic subunit (ClpP) is an enzyme that in humans is encoded by the CLPP gene. This protein is an essential component to form the protein complex of Clp protease.
Intermediate cleaving peptidase 55 is an enzyme. This enzyme catalyses the following chemical reaction
Signal peptidase I is an enzyme. This enzyme catalyses the following chemical reaction
Mitochondrial processing peptidase is an enzyme complex found in mitochondria which cleaves signal sequences from mitochondrial proteins. In humans this complex is composed of two subunits encoded by the genes PMPCA, and PMPCB. The enzyme is also known as. This enzyme catalyses the following chemical reaction

Mitochondrial intermediate peptidase is an enzyme that in humans is encoded by the MIPEP gene. This protein is a critical component of human mitochondrial protein import machinery involved in the maturing process of nuclear coded mitochondrial proteins that with a mitochondrial translocation peptide, especially those OXPHOS-related proteins.

Inner mitochondrial membrane peptidase subunit 2 (IMMP2L) is an enzyme that in humans is encoded by the IMMP2L gene on chromosome 7. This protein catalyzes the removal of transit peptides required for the targeting of proteins from the mitochondrial matrix, across the inner membrane, into the intermembrane space. IMMP2L processes the nuclear encoded protein DIABLO.
References
- ↑ Isaya G, Kalousek F, Rosenberg LE (April 1992). "Amino-terminal octapeptides function as recognition signals for the mitochondrial intermediate peptidase". The Journal of Biological Chemistry. 267 (11): 7904–10. doi: 10.1016/S0021-9258(18)42598-7 . PMID 1560019.
- ↑ Isaya G, Kalousek F, Rosenberg LE (September 1992). "Sequence analysis of rat mitochondrial intermediate peptidase: similarity to zinc metallopeptidases and to a putative yeast homologue". Proceedings of the National Academy of Sciences of the United States of America. 89 (17): 8317–21. doi: 10.1073/pnas.89.17.8317 . PMC 49909 . PMID 1518864.