
Xenopus is a genus of highly aquatic frogs native to sub-Saharan Africa. Twenty species are currently described within it. The two best-known species of this genus are Xenopus laevis and Xenopus tropicalis, which are commonly studied as model organisms for developmental biology, cell biology, toxicology, neuroscience and for modelling human disease and birth defects.
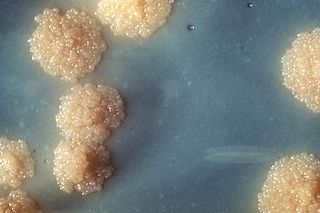
Mycobacterium tuberculosis is a species of pathogenic bacteria in the family Mycobacteriaceae and the causative agent of tuberculosis. First discovered in 1882 by Robert Koch, M. tuberculosis has an unusual, waxy coating on its cell surface primarily due to the presence of mycolic acid. This coating makes the cells impervious to Gram staining, and as a result, M. tuberculosis can appear either Gram-negative or Gram-positive. Acid-fast stains such as Ziehl-Neelsen, or fluorescent stains such as auramine are used instead to identify M. tuberculosis with a microscope. The physiology of M. tuberculosis is highly aerobic and requires high levels of oxygen. Primarily a pathogen of the mammalian respiratory system, it infects the lungs. The most frequently used diagnostic methods for tuberculosis are the tuberculin skin test, acid-fast stain, culture, and polymerase chain reaction.
Nontuberculous mycobacteria (NTM), also known as environmental mycobacteria, atypical mycobacteria and mycobacteria other than tuberculosis (MOTT), are mycobacteria which do not cause tuberculosis or leprosy. NTM do cause pulmonary diseases that resemble tuberculosis. Mycobacteriosis is any of these illnesses, usually meant to exclude tuberculosis. They occur in many animals, including humans.

Buruli ulcer is an infectious disease caused by Mycobacterium ulcerans. The early stage of the infection is characterised by a painless nodule or area of swelling. This nodule can turn into an ulcer. The ulcer may be larger inside than at the surface of the skin, and can be surrounded by swelling. As the disease worsens, bone can be infected. Buruli ulcers most commonly affect the arms or legs; fever is uncommon.
The rpoB gene encodes the β subunit of bacterial RNA polymerase. rpoB is also found in plant chloroplasts where it forms the beta subunit of the plastid-encoded RNA polymerase (PEP). An inhibitor of transcription in bacteria, tagetitoxin, also inhibits PEP, showing that the complex found in plants is very similar to the homologous enzyme in bacteria. It codes for 1342 amino acids, making it the second-largest polypeptide in the bacterial cell. It is the site of mutations that confer resistance to the rifamycin antibacterial agents, such as rifampin. Mutations in rpoB that confer resistance to rifamycins do so by altering residues of the rifamycin binding site on RNA polymerase, thereby reducing rifamycin binding affinity for rifamycins
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.
Mycobacterium pseudoshottsii, a slowly growing chromogenic species was isolated from Chesapeake Bay striped bass during an epizootic of mycobacteriosis.
Mycobacterium shottsii is a slowly growing, non-pigmented mycobacteria isolated from striped bass during an epizootic of mycobacteriosis in the Chesapeake Bay. Growth characteristics, acid-fastness and results of 16S rRNA gene sequencing were consistent with those of the genus Mycobacterium. A unique profile of biochemical reactions was observed among the 21 isolates. A single cluster of eight peaks identified by analysis of mycolic acids (HPLC) resembled those of reference patterns but differed in peak elution times from profiles of reference species of the Mycobacterium tuberculosis complex. Etymology: shottsii; of Shotts, named after Emmett Shotts, an American fish bacteriologist.
The Runyon classification of nontuberculous mycobacteria based on the rate of growth, production of yellow pigment and whether this pigment was produced in the dark or only after exposure to light.
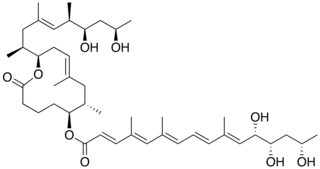
Mycolactone is a polyketide-derived macrolide produced and secreted by a group of very closely related pathogenic Mycobacteria species that have been assigned a variety of names including, M. ulcerans, M. liflandii, M. pseudoshottsii, and some strains of M. marinum. These mycobacteria are collectively referred to as mycolactone-producing mycobacteria or MPM.
Mycobacterium abscessus complex (MABSC) is a group of rapidly growing, multidrug-resistant non-tuberculous mycobacteria (NTM) species that are common soil and water contaminants. Although M. abscessus complex most commonly cause chronic lung infection and skin and soft tissue infection (SSTI), the complex can also cause infection in almost all human organs, mostly in patients with suppressed immune systems. Amongst NTM species responsible for disease, infection caused by M. abscessus complex are more difficult to treat due to antimicrobial drug resistance.
Mycobacterium conceptionense is a non pigmented rapidly growing mycobacterium was first isolated from wound liquid outflow, bone tissue biopsy, and excised skin tissue from a 31-year-old woman who suffered an accidental open right tibia fracture and prolonged stay in a river. Etymology: conceptionense, pertaining to Hôpital de la Conception, the hospital where the first strain was isolated.

Mycobacterium cosmeticum is a rapidly growing mycobacterium that was first isolated from cosmetic patients and sites performing cosmetic procedures.
Mycobacterium massiliense is a rapidly growing Mycobacteria species sharing an identical 16S rRNA sequence with Mycobacterium abscessus. Etymology: massiliense, pertaining to Massilia, the Latin name of Marseille, France where the organism was isolated.
Mycobacterium montefiorense is a species of bacteria which cause granulomatous skin disease of moray eels. Sequence analysis, of the 16S rRNA gene reveals M. montefiorense is most closely related to Mycobacterium triplex, an opportunistic pathogen of humans.

DsrA RNA is a non-coding RNA that regulates both transcription, by overcoming transcriptional silencing by the nucleoid-associated H-NS protein, and translation, by promoting efficient translation of the stress sigma factor, RpoS. These two activities of DsrA can be separated by mutation: the first of three stem-loops of the 85 nucleotide RNA is necessary for RpoS translation but not for anti-H-NS action, while the second stem-loop is essential for antisilencing and less critical for RpoS translation. The third stem-loop, which behaves as a transcription terminator, can be substituted by the trp transcription terminator without loss of either DsrA function. The sequence of the first stem-loop of DsrA is complementary with the upstream leader portion of RpoS messenger RNA, suggesting that pairing of DsrA with the RpoS message might be important for translational regulation. The structures of DsrA and DsrA/rpoS complex were studied by NMR. The study concluded that the sRNA contains a dynamic conformational equilibrium for its second stem–loop which might be an important mechanism for DsrA to regulate the translations of its multiple target mRNAs.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.







