Related Research Articles

Pasteurellosis is an infection with a species of the bacterial genus Pasteurella, which is found in humans and other animals.

Concanavalin A (ConA) is a lectin originally extracted from the jack-bean. It is a member of the legume lectin family. It binds specifically to certain structures found in various sugars, glycoproteins, and glycolipids, mainly internal and nonreducing terminal α-D-mannosyl and α-D-glucosyl groups. Its physiological function in plants, however, is still unknown. ConA is a plant mitogen, and is known for its ability to stimulate mouse T-cell subsets giving rise to four functionally distinct T cell populations, including precursors to regulatory T cells; a subset of human suppressor T-cells is also sensitive to ConA. ConA was the first lectin to be available on a commercial basis, and is widely used in biology and biochemistry to characterize glycoproteins and other sugar-containing entities on the surface of various cells. It is also used to purify glycosylated macromolecules in lectin affinity chromatography, as well as to study immune regulation by various immune cells.
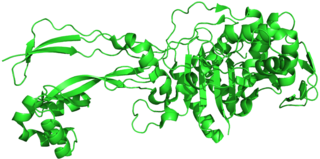
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.

Furin is a protease, a proteolytic enzyme that in humans and other animals is encoded by the FURIN gene. Some proteins are inactive when they are first synthesized, and must have sections removed in order to become active. Furin cleaves these sections and activates the proteins. It was named furin because it was in the upstream region of an oncogene known as FES. The gene was known as FUR and therefore the protein was named furin. Furin is also known as PACE. A member of family S8, furin is a subtilisin-like peptidase.
The indole test is a biochemical test performed on bacterial species to determine the ability of the organism to convert tryptophan into indole. This division is performed by a chain of a number of different intracellular enzymes, a system generally referred to as "tryptophanase."

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.

Lysins, also known as endolysins or murein hydrolases, are hydrolytic enzymes produced by bacteriophages in order to cleave the host's cell wall during the final stage of the lytic cycle. Lysins are highly evolved enzymes that are able to target one of the five bonds in peptidoglycan (murein), the main component of bacterial cell walls, which allows the release of progeny virions from the lysed cell. Cell-wall-containing Archaea are also lysed by specialized pseudomurein-cleaving lysins, while most archaeal viruses employ alternative mechanisms. Similarly, not all bacteriophages synthesize lysins: some small single-stranded DNA and RNA phages produce membrane proteins that activate the host's autolytic mechanisms such as autolysins.
Kexin is a prohormone-processing protease, specifically a yeast serine peptidase, found in the budding yeast. It catalyzes the cleavage of -Lys-Arg- and -Arg-Arg- bonds to process yeast alpha-factor pheromone and killer toxin precursors. The human homolog is PCSK4. It is a family of subtilisin-like peptidases. Even though there are a few prokaryote kexin-like peptidases, all kexins are eukaryotes. The enzyme is encoded by the yeast gene KEX2, and usually referred to in the scientific community as Kex2p. It shares structural similarities with the bacterial protease subtilisin. The first mammalian homologue of this protein to be identified was furin. In the mammal, kexin-like peptidases function in creating and regulating many differing proproteins.

Glycophorin A (MNS blood group), also known as GYPA, is a protein which in humans is encoded by the GYPA gene. GYPA has also recently been designated CD235a (cluster of differentiation 235a).

Glycophorin B (MNS blood group) (gene designation GYPB) also known as sialoglycoprotein delta and SS-active sialoglycoprotein is a protein which in humans is encoded by the GYPB gene. GYPB has also recently been designated CD235b (cluster of differentiation 235b).

Probable O-sialoglycoprotein endopeptidase is an enzyme that in humans is encoded by the OSGEP gene.
Feline coronavirus (FCoV) is a positive-stranded RNA virus that infects cats worldwide. It is a coronavirus of the species Alphacoronavirus 1 which includes canine coronavirus (CCoV) and porcine transmissible gastroenteritis coronavirus (TGEV). It has two different forms: feline enteric coronavirus (FECV) that infects the intestines and feline infectious peritonitis virus (FIPV) that causes the disease feline infectious peritonitis (FIP).
IgA protease is an enzyme. This enzyme catalyses the following chemical reaction[reaction equation needed]

Glutamyl endopeptidase is an extracellular bacterial serine protease of the glutamyl endopeptidase I family that was initially isolated from the Staphylococcus aureus strain V8. The protease is, hence, commonly referred to as "V8 protease", or alternatively SspA from its corresponding gene.
Leishmanolysin is an enzyme. This enzyme catalyses the following chemical reaction
Oligopeptidase A is an enzyme. This enzyme catalyses the following chemical reaction
Mannheimia ruminalis is a bacterium. It is pathogenic.
Mannheimia varigena is a bacterium, predominantly encountered in ruminants and historically classified within the former bacterial Pasteurella haemolytica complex, a group of bacteria involved in bovine respiratory disease (BRD). It is pathogenic.
Bovine respiratory disease (BRD) is the most common and costly disease affecting beef cattle in the world. It is a complex, bacterial or viral infection that causes pneumonia in calves which can be fatal. The infection is usually a sum of three codependent factors: stress, an underlying viral infection, and a new bacterial infection. The diagnosis of the disease is complex since there are multiple possible causes.
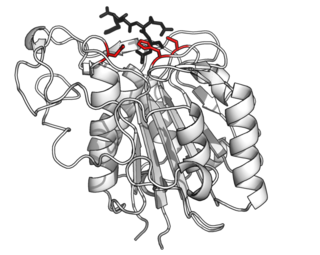
Asparagine endopeptidase is a proteolytic enzyme from C13 peptidase family which hydrolyses a peptide bond using the thiol group of a cysteine residue as a nucleophile. It is also known as asparaginyl endopeptidase, citvac, proteinase B, hemoglobinase, PRSC1 gene product or LGMN, vicilin peptidohydrolase and bean endopeptidase. In humans it is encoded by the LGMN gene.
References
- ↑ Abdullah KM, Lo RY, Mellors A (September 1991). "Cloning, nucleotide sequence, and expression of the Pasteurella haemolytica A1 glycoprotease gene". Journal of Bacteriology. 173 (18): 5597–603. doi:10.1128/jb.173.18.5597-5603.1991. PMC 208286 . PMID 1885539.
- ↑ Abdullah KM, Udoh EA, Shewen PE, Mellors A (January 1992). "A neutral glycoprotease of Pasteurella haemolytica A1 specifically cleaves O-sialoglycoproteins". Infection and Immunity. 60 (1): 56–62. PMC 257502 . PMID 1729196.
- ↑ Sutherland DR, Abdullah KM, Cyopick P, Mellors A (March 1992). "Cleavage of the cell-surface O-sialoglycoproteins CD34, CD43, CD44, and CD45 by a novel glycoprotease from Pasteurella haemolytica". Journal of Immunology. 148 (5): 1458–64. PMID 1371528.