Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.

Muconate lactonizing enzymes are involved in the breakdown of lignin-derived aromatics, catechol and protocatechuate, to citric acid cycle intermediates as a part of the β-ketoadipate pathway in soil microbes. Some bacterial species are also capable of dehalogenating chloroaromatic compounds by the action of chloromuconate lactonizing enzymes. MLEs consist of several strands which have variable reaction favorable parts therefore the configuration of the strands affect its ability to accept protons. The bacterial MLEs belong to the enolase superfamily, several structures from which are known. MLEs have an identifying structure made up of two proteins and two Magnesium ions as well as various classes depending on whether it is bacterial or eukaryotic. The reaction mechanism that MLEs undergo are the reverse of beta-elimination in which the enolate alpha-carbon is protonated. MLEs can undergo mutations caused by a deletion of catB structural genes which can cause some bacteria to lose its functions such as the ability to grow. Additional mutations to MLEs can cause its structure and function to alter and could cause the conformation to change therefore making it an inactive enzyme that is unable to bind its substrate. There is another enzyme called Mandelate Racemase that is very similar to MLEs in the structural way as well as them both being a part of the enolase superfamily. They both have the same end product even though they undergo different chemical reactions in order to reach the end product.

Orotidine 5'-phosphate decarboxylase or orotidylate decarboxylase is an enzyme involved in pyrimidine biosynthesis. It catalyzes the decarboxylation of orotidine monophosphate (OMP) to form uridine monophosphate (UMP). The function of this enzyme is essential to the de novo biosynthesis of the pyrimidine nucleotides uridine triphosphate, cytidine triphosphate, and thymidine triphosphate. OMP decarboxylase has been a frequent target for scientific investigation because of its demonstrated extreme catalytic efficiency and its usefulness as a selection marker for yeast strain engineering.
The crotonase family comprises mechanistically diverse proteins that share a conserved trimeric quaternary structure, the core of which consists of 4 turns of a (beta/beta/alpha)n superhelix.

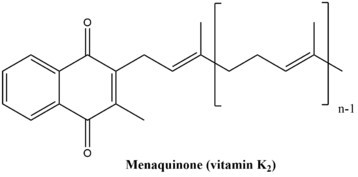
Isochorismate synthase ( EC 5.4.4.2) is an isomerase enzyme that catalyzes the first step in the biosynthesis of vitamin K2 (menaquinone) in Escherichia coli.

The enzyme UDP-glucose 4-epimerase, also known as UDP-galactose 4-epimerase or GALE, is a homodimeric epimerase found in bacterial, fungal, plant, and mammalian cells. This enzyme performs the final step in the Leloir pathway of galactose metabolism, catalyzing the reversible conversion of UDP-galactose to UDP-glucose. GALE tightly binds nicotinamide adenine dinucleotide (NAD+), a co-factor required for catalytic activity.

The enzyme chorismate lyase catalyzes the chemical reaction

The enzyme 1,4-dihydroxy-2-naphthoyl-CoA synthase catalyzes the sixth step in the biosynthesis of phylloquinone and menaquinone, the two forms of vitamin K. In E. coli, 1,4-dihydroxy-2-naphthoyl-CoA synthase, formerly known as naphthoate synthase, is encoded by menB and uses O-succinylbenzoyl-CoA as a substrate and converts it to 1,4-dihydroxy-2-naphthoyl-CoA.
In enzymology, a 5-(carboxyamino)imidazole ribonucleotide synthase (EC 6.3.4.18) is an enzyme that catalyzes the chemical reaction

In enzymology, an aminodeoxychorismate synthase is an enzyme that catalyzes the chemical reaction

o-Succinylbenzoate—CoA ligase, encoded from the menE gene in Escherichia coli, catalyzes the fifth reaction in the synthesis of menaquinone. This pathway is called 1, 4-dihydroxy-2-naphthoate biosynthesis I. Vitamin K is a quinone that serves as an electron transporter during anaerobic respiration. This process of anaerobic respiration allows the bacteria to generate the energy required to survive.

In molecular biology, the protein domain SAICAR synthase is an enzyme which catalyses a reaction to create SAICAR. In enzymology, this enzyme is also known as phosphoribosylaminoimidazolesuccinocarboxamide synthase. It is an enzyme that catalyzes the chemical reaction
In enzymology, a chorismate synthase is an enzyme that catalyzes the chemical reaction
In enzymology, a [acyl-carrier-protein] S-malonyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a malate synthase (EC 2.3.3.9) is an enzyme that catalyzes the chemical reaction
2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, also known as SHCHC synthase, is encoded by the menH gene in E.coli and functions in the synthesis of vitamin K. The specific step in the synthetic pathway that SHCHC synthase catalyzes is the conversion of 5-enolpyruvoyl-6-hydroxy-2-succinylcyclohex-3-ene-1-carboxylate to (1R,6R)-6-hydroxy-2-succinylcyclohexa-2,4-diene-1-carboxylate and pyruvate.

The Enzyme Function Initiative (EFI) is a large-scale collaborative project aiming to develop and disseminate a robust strategy to determine enzyme function through an integrated sequence–structure-based approach. The project was funded in May 2010 by the National Institute of General Medical Sciences as a Glue Grant which supports the research of complex biological problems that cannot be solved by a single research group. The EFI was largely spurred by the need to develop methods to identify the functions of the enormous number proteins discovered through genomic sequencing projects.
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical, as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.
1-4-dihydroxy-2-napthoate (DHNA) polyprenyltransferase (EC 2.5.1.74)is an enzyme that catalyzes the chemical reaction: all-trans-nonaprenyl diphosphate + 1-4-dihydroxy-2-napthoate + H+ demethylmenaquinol-9 + diphosphate + carbon dioxide
Glycerate 2-kinase is an enzyme with systematic name ATP:D-glycerate 2-phosphotransferase. This enzyme catalyses the following chemical reaction