Related Research Articles
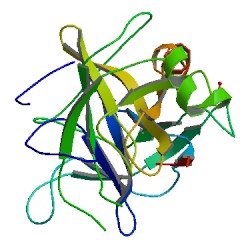
Chymotrypsin (EC 3.4.21.1, chymotrypsins A and B, alpha-chymar ophth, avazyme, chymar, chymotest, enzeon, quimar, quimotrase, alpha-chymar, alpha-chymotrypsin A, alpha-chymotrypsin) is a digestive enzyme component of pancreatic juice acting in the duodenum, where it performs proteolysis, the breakdown of proteins and polypeptides. Chymotrypsin preferentially cleaves peptide amide bonds where the side chain of the amino acid N-terminal to the scissile amide bond (the P1 position) is a large hydrophobic amino acid (tyrosine, tryptophan, and phenylalanine). These amino acids contain an aromatic ring in their side chain that fits into a hydrophobic pocket (the S1 position) of the enzyme. It is activated in the presence of trypsin. The hydrophobic and shape complementarity between the peptide substrate P1 side chain and the enzyme S1 binding cavity accounts for the substrate specificity of this enzyme. Chymotrypsin also hydrolyzes other amide bonds in peptides at slower rates, particularly those containing leucine at the P1 position.

Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion.

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
Aspergillopepsin I is an enzyme. This enzyme catalyses the following chemical reaction
In molecular biology, the Signal Peptide Peptidase (SPP) is a type of protein that specifically cleaves parts of other proteins. It is an intramembrane aspartyl protease with the conserved active site motifs 'YD' and 'GxGD' in adjacent transmembrane domains (TMDs). Its sequences is highly conserved in different vertebrate species. SPP cleaves remnant signal peptides left behind in membrane by the action of signal peptidase and also plays key roles in immune surveillance and the maturation of certain viral proteins.

In molecular biology, Proteinase K is a broad-spectrum serine protease. The enzyme was discovered in 1974 in extracts of the fungus Parengyodontium album. Proteinase K is able to digest hair (keratin), hence, the name "Proteinase K". The predominant site of cleavage is the peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups. It is commonly used for its broad specificity. This enzyme belongs to Peptidase family S8 (subtilisin). The molecular weight of Proteinase K is 28,900 daltons.

Cathepsin E is an enzyme that in humans is encoded by the CTSE gene. The enzyme is also known as slow-moving proteinase, erythrocyte membrane aspartic proteinase, SMP, EMAP, non-pepsin proteinase, cathepsin D-like acid proteinase, cathepsin E-like acid proteinase, cathepsin D-type proteinase) is an enzyme.
Nepenthesin is an aspartic protease of plant origin that has so far been identified in the pitcher secretions of Nepenthes and in the leaves of Drosera peltata. It is similar to pepsin, but differs in that it also cleaves on either side of Asp residues and at Lys┼Arg. While more pH and temperature stable than porcine pepsin A, it is considerably less stable in urea or guanidine hydrochloride. It is the only known protein with such a stability profile.
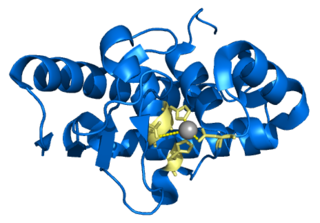
Lys-N is a metalloendopeptidase found in the mushroom Grifola frondosa that cleaves proteins on the amino side of lysine residues.

Cyanophycinase (EC 3.4.15.6, cyanophycin degrading enzyme, beta-Asp-Arg hydrolysing enzyme, CGPase, CphB, CphE, cyanophycin granule polypeptidase, extracellular CGPase) is an enzyme. It catalyses the following chemical reaction
Lysyl endopeptidase is an enzyme. This enzyme catalyses the following chemical reaction
Caricain is an enzyme. This enzyme catalyses the following chemical reaction: Hydrolysis of proteins with broad specificity for peptide bonds, similar to those of papain and chymopapain
Adenain is an enzyme. This enzyme catalyses the following chemical reaction
Candidapepsin is an enzyme. This enzyme catalyses the following chemical reaction
Peptidyl-Lys metalloendopeptidase is an enzyme. This enzyme catalyses the following chemical reaction
Beta-lytic metalloendopeptidase is an enzyme. This enzyme catalyses the following chemical reaction
Mitochondrial processing peptidase is an enzyme complex found in mitochondria which cleaves signal sequences from mitochondrial proteins. In humans this complex is composed of two subunits encoded by the genes PMPCA, and PMPCB. The enzyme is also known as. This enzyme catalyses the following chemical reaction
Asparagine peptide lyase are one of the seven groups in which proteases, also termed proteolytic enzymes, peptidases, or proteinases, are classified according to their catalytic residue. The catalytic mechanism of the asparagine peptide lyases involves an asparagine residue acting as nucleophile to perform a nucleophilic elimination reaction, rather than hydrolysis, to catalyse the breaking of a peptide bond.

The sedolisin family of peptidases are a family of serine proteases structurally related to the subtilisin (S8) family. Well-known members of this family include sedolisin ("pseudomonalisin") found in Pseudomonas bacteria, xanthomonalisin ("sedolisin-B"), physarolisin as well as animal tripeptidyl peptidase I. It is also known as sedolysin or serine-carboxyl peptidase. This group of enzymes contains a variation on the catalytic triad: unlike S8 which uses Ser-His-Asp, this group runs on Ser-Glu-Asp, with an additional acidic residue Asp in the oxyanion hole.
References
- ↑ Porzio MA, Pearson AM (March 1975). "Isolation of an extracellular neutral proteinase from Pseudomonas fragi". Biochimica et Biophysica Acta (BBA) - Enzymology. 384 (1): 235–241. doi:10.1016/0005-2744(75)90112-6. PMID 236771.
- ↑ Drapeau GR (February 1980). "Substrate specificity of a proteolytic enzyme isolated from a mutant of Pseudomonas fragi". The Journal of Biological Chemistry. 255 (3): 839–840. doi: 10.1016/S0021-9258(19)86106-9 . PMID 7188696.
- ↑ Ingrosso D, Fowler AV, Bleibaum J, Clarke S (August 1989). "Specificity of endoproteinase Asp-N (Pseudomonas fragi): cleavage at glutamyl residues in two proteins". Biochemical and Biophysical Research Communications. 162 (3): 1528–1534. doi:10.1016/0006-291x(89)90848-6. PMID 2669754.
- ↑ Noreau J, Drapeau GR (December 1979). "Isolation and properties of the protease from the wild-type and mutant strains of Pseudomonas fragi". Journal of Bacteriology. 140 (3): 911–916. doi:10.1128/JB.140.3.911-916.1979. PMC 216733 . PMID 42639.
- ↑ Hagmann ML, Geuss U, Fischer S, Kresse GB (1995). Peptidyl-Asp metalloendopeptidase. Methods in Enzymology. Vol. 248. pp. 782–787. doi:10.1016/0076-6879(95)48053-6. PMID 7674963.
- ↑ Ellard JM, Zollitsch T, Cummins WJ, Hamilton AL, Bradley M (September 2002). "Fluorescence enhancement through enzymatic cleavage of internally quenched dendritic peptides: a sensitive assay for the AspN endoproteinase". Angewandte Chemie. 41 (17): 3233–3236. doi:10.1002/1521-3773(20020902)41:17<3233::AID-ANIE3233>3.0.CO;2-E. PMID 12207399.
- ↑ Dai S, Lee DH, Kenton NT, Zhou ZS (2013). Handbook of Proteolytic Enzymes. Oxford: Academic Press. pp. 1281–1285. ISBN 978-0-12-382219-2.
- ↑ Ni W, Dai S, Karger BL, Zhou ZS (September 2010). "Analysis of isoaspartic Acid by selective proteolysis with Asp-N and electron transfer dissociation mass spectrometry". Analytical Chemistry. 82 (17): 7485–7491. doi:10.1021/ac101806e. PMC 2936825 . PMID 20712325.