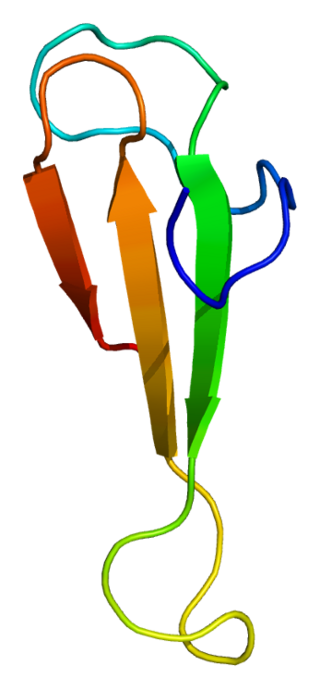Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA, while at least 80% of mammalian genomic DNA can be actively transcribed, with the majority of this 80% considered to be ncRNA.

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.
A sigma factor is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP).
In molecular biology, the TATA box is a sequence of DNA found in the core promoter region of genes in archaea and eukaryotes. The bacterial homolog of the TATA box is called the Pribnow box which has a shorter consensus sequence.

The preinitiation complex is a complex of approximately 100 proteins that is necessary for the transcription of protein-coding genes in eukaryotes and archaea. The preinitiation complex positions RNA polymerase II at gene transcription start sites, denatures the DNA, and positions the DNA in the RNA polymerase II active site for transcription.

RNA polymerase II is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription.

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.
In eukaryote cells, RNA polymerase III is a protein that transcribes DNA to synthesize 5S ribosomal RNA, tRNA and other small RNAs.

The TATA-binding protein (TBP) is a general transcription factor that binds specifically to a DNA sequence called the TATA box. This DNA sequence is found about 30 base pairs upstream of the transcription start site in some eukaryotic gene promoters.
Transcription factor II D (TFIID) is one of several general transcription factors that make up the RNA polymerase II preinitiation complex. RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins. Before the start of transcription, the transcription Factor II D (TFIID) complex binds to the core promoter DNA of the gene through specific recognition of promoter sequence motifs, including the TATA box, Initiator, Downstream Promoter, Motif Ten, or Downstream Regulatory elements.
Antitermination is the prokaryotic cell's aid to fix premature termination of RNA synthesis during the transcription of RNA. It occurs when the RNA polymerase ignores the termination signal and continues elongating its transcript until a second signal is reached. Antitermination provides a mechanism whereby one or more genes at the end of an operon can be switched either on or off, depending on the polymerase either recognizing or not recognizing the termination signal.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.

Intrinsic, or rho-independent termination, is a process in prokaryotes to signal the end of transcription and release the newly constructed RNA molecule. In prokaryotes such as E. coli, transcription is terminated either by a rho-dependent process or rho-independent process. In the Rho-dependent process, the rho-protein locates and binds the signal sequence in the mRNA and signals for cleavage. Contrarily, intrinsic termination does not require a special protein to signal for termination and is controlled by the specific sequences of RNA. When the termination process begins, the transcribed mRNA forms a stable secondary structure hairpin loop, also known as a Stem-loop. This RNA hairpin is followed by multiple uracil nucleotides. The bonds between uracil and adenine are very weak. A protein bound to RNA polymerase (nusA) binds to the stem-loop structure tightly enough to cause the polymerase to temporarily stall. This pausing of the polymerase coincides with transcription of the poly-uracil sequence. The weak adenine-uracil bonds lower the energy of destabilization for the RNA-DNA duplex, allowing it to unwind and dissociate from the RNA polymerase. Overall, the modified RNA structure is what terminates transcription.
Transcription factor II B (TFIIB) is a general transcription factor that is involved in the formation of the RNA polymerase II preinitiation complex (PIC) and aids in stimulating transcription initiation. TFIIB is localised to the nucleus and provides a platform for PIC formation by binding and stabilising the DNA-TBP complex and by recruiting RNA polymerase II and other transcription factors. It is encoded by the TFIIB gene, and is homologous to archaeal transcription factor B and analogous to bacterial sigma factors.
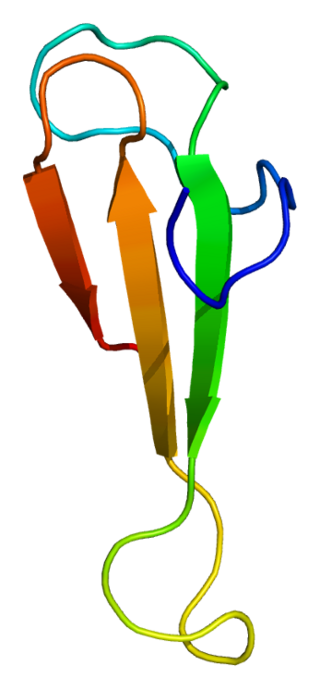
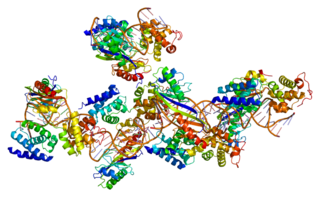
Transcription elongation factor A protein 1 is a protein that in humans is encoded by the TCEA1 gene.
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.
In gene expression, DSIF is a protein that can either negatively or positively affect transcription by RNA polymerase II. In one case of negative regulation, it can interact with negative elongation factor (NELF) to promote the stalling of Pol II at some genes. This stalling is relieved by P-TEFb. In humans, DSIF is composed of hSPT4 and hSPT5.
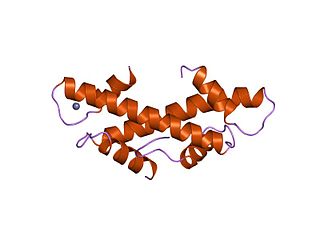
The TBP-associated factors (TAF) are proteins that associate with the TATA-binding protein in transcription initiation. It is a part of the transcription initiation factor TFIID multimeric protein complex. It also makes up many other factors, including SL1. They mediate the formation of the transcription preinitiation complex, a step preceding transcription of DNA to RNA by RNA polymerase II.

Archaeal transcription factor B is a protein family of extrinsic transcription factors that guide the initiation of RNA transcription in organisms that fall under the domain of Archaea. It is homologous to eukaryotic TFIIB and, more distantly, to bacterial sigma factor. Like these proteins, it is involved in forming transcription preinitiation complexes. Its structure includes several conserved motifs which interact with DNA and other transcription factors, notably the single type of RNA polymerase that performs transcription in Archaea.