
In stereochemistry, stereoisomerism, or spatial isomerism, is a form of isomerism in which molecules have the same molecular formula and sequence of bonded atoms (constitution), but differ in the three-dimensional orientations of their atoms in space. This contrasts with structural isomers, which share the same molecular formula, but the bond connections or their order differs. By definition, molecules that are stereoisomers of each other represent the same structural isomer.
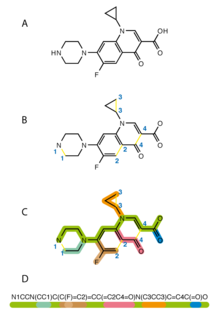
The simplified molecular-input line-entry system (SMILES) is a specification in the form of a line notation for describing the structure of chemical species using short ASCII strings. SMILES strings can be imported by most molecule editors for conversion back into two-dimensional drawings or three-dimensional models of the molecules.

The structural formula of a chemical compound is a graphic representation of the molecular structure, showing how the atoms are possibly arranged in the real three-dimensional space. The chemical bonding within the molecule is also shown, either explicitly or implicitly. Unlike chemical formulas, which have a limited number of symbols and are capable of only limited descriptive power, structural formulas provide a more complete geometric representation of the molecular structure. For example, many chemical compounds exist in different isomeric forms, which have different enantiomeric structures but the same chemical formula.
Irrlicht is an open-source game engine written in C++. It is cross-platform, officially running on Windows, macOS, Linux and Windows CE and due to its open nature ports to other systems are available, including FreeBSD, Xbox, PlayStation Portable, Symbian, iPhone, AmigaOS 4 and Google Native Client.
The skeletal formula, also called line-angle formula or shorthand formula, of an organic compound is a type of molecular structural formula that serves as a shorthand representation of a molecule's bonding and some details of its molecular geometry. A skeletal formula shows the skeletal structure or skeleton of a molecule, which is composed of the skeletal atoms that make up the molecule. It is represented in two dimensions, as on a piece of paper. It employs certain conventions to represent carbon and hydrogen atoms, which are the most common in organic chemistry.
Cairo is an open source graphics library that provides a vector graphics-based, device-independent API for software developers. It provides primitives for two-dimensional drawing across a number of different back ends. Cairo uses hardware acceleration when available.
This article discusses some common molecular file formats, including usage and converting between them.
Chemical table file is a family of text-based chemical file formats that describe molecules and chemical reactions. One format, for example, lists each atom in a molecule, the x-y-z coordinates of that atom, and the bonds among the atoms.
The IUPAC International Chemical Identifier is a textual identifier for chemical substances, designed to provide a standard way to encode molecular information and to facilitate the search for such information in databases and on the web. Initially developed by IUPAC and NIST from 2000 to 2005, the format and algorithms are non-proprietary.

PyMOL is an open source molecular visualization system created by Warren Lyford DeLano. It was commercialized initially by DeLano Scientific LLC, which was a private software company dedicated to creating useful tools that become universally accessible to scientific and educational communities. It is currently commercialized by Schrödinger, Inc. PyMOL can produce high-quality 3D images of small molecules and biological macromolecules, such as proteins. According to the original author, by 2009, almost a quarter of all published images of 3D protein structures in the scientific literature were made using PyMOL.

XDrawChem is a free software program for drawing chemical structural formulas, available for Unix and macOS. It is distributed under the GNU GPL. In Microsoft Windows this program is called WinDrawChem.

Jmol is computer software for molecular modelling chemical structures in 3-dimensions. Jmol returns a 3D representation of a molecule that may be used as a teaching tool, or for research e.g., in chemistry and biochemistry. It is written in the programming language Java, so it can run on the operating systems Windows, macOS, Linux, and Unix, if Java is installed. It is free and open-source software released under a GNU Lesser General Public License (LGPL) version 2.0. A standalone application and a software development kit (SDK) exist that can be integrated into other Java applications, such as Bioclipse and Taverna.

JOELib is computer software, a chemical expert system used mainly to interconvert chemical file formats. Because of its strong relationship to informatics, this program belongs more to the category cheminformatics than to molecular modelling. It is available for Windows, Unix and other operating systems supporting the programming language Java. It is free and open-source software distributed under the GNU General Public License (GPL) 2.0.

The Chemistry Development Kit (CDK) is computer software, a library in the programming language Java, for chemoinformatics and bioinformatics. It is available for Windows, Linux, Unix, and macOS. It is free and open-source software distributed under the GNU Lesser General Public License (LGPL) 2.0.

ISIS/Draw was a chemical structure drawing program developed by MDL Information Systems. It introduced a number of file formats for the storage of chemical information that have become industry standards.

BALL is software consisting of the versatile C++ class framework Biochemical Algorithms Library (BALL), a set of algorithms and data structures for molecular modelling and computational structural bioinformatics, a Python interface to this library, and a graphical user interface to BALL, the molecule viewer BALLView.
The SYBYL line notation or SLN is a specification for unambiguously describing the structure of chemical molecules using short ASCII strings. SLN differs from SMILES in several significant ways. SLN can specify molecules, molecular queries, and reactions in a single line notation whereas SMILES handles these through language extensions. SLN has support for relative stereochemistry, it can distinguish mixtures of enantiomers from pure molecules with pure but unresolved stereochemistry. In SMILES aromaticity is considered to be a property of both atoms and bonds whereas in SLN it is a property of bonds.

The JME Molecule Editor is a molecule editor Java applet with which users make and edit drawings of molecules and reactions, and can display molecules within an HTML page. The editor can generate Daylight simplified molecular-input line-entry system (SMILES) or MDL Molfiles of the created structures.

ChemSpider is a database of chemicals. ChemSpider is owned by the Royal Society of Chemistry.

Tryton is a three-tier high-level general purpose computer application platform on top of which is built an Enterprise resource planning (ERP) business solution through a set of Tryton modules. The three-tier architecture consists of the Tryton client, the Tryton server and the Database management system.
















