
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequence—that is, the prediction of its secondary and tertiary structure from primary structure. Structure prediction is different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by computational biology; it is important in medicine and biotechnology.
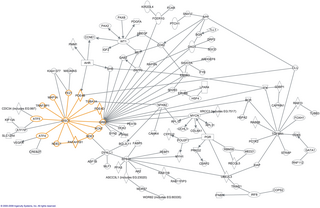
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules but can also describe sets of indirect interactions among genes.

Michael Levitt, is a South African-born biophysicist and a professor of structural biology at Stanford University, a position he has held since 1987. Levitt received the 2013 Nobel Prize in Chemistry, together with Martin Karplus and Arieh Warshel, for "the development of multiscale models for complex chemical systems". In 2018, Levitt was a founding co-editor of the Annual Review of Biomedical Data Science.

Intelligent Systems for Molecular Biology (ISMB) is an annual academic conference on the subjects of bioinformatics and computational biology organised by the International Society for Computational Biology (ISCB). The principal focus of the conference is on the development and application of advanced computational methods for biological problems. The conference has been held every year since 1993 and has grown to become one of the largest and most prestigious meetings in these fields, hosting over 2,000 delegates in 2004. From the first meeting, ISMB has been held in locations worldwide; since 2007, meetings have been located in Europe and North America in alternating years. Since 2004, European meetings have been held jointly with the European Conference on Computational Biology (ECCB).
Anders Krogh is a bioinformatician at the University of Copenhagen, where he leads the university's bioinformatics center. He is known for his pioneering work on the use of hidden Markov models in bioinformatics, and is co-author of a widely used textbook in bioinformatics. In addition, he also co-authored one of the early textbooks on neural networks. His current research interests include promoter analysis, non-coding RNA, gene prediction and protein structure prediction.
Orientations of Proteins in Membranes (OPM) database provides spatial positions of membrane protein structures with respect to the lipid bilayer. Positions of the proteins are calculated using an implicit solvation model of the lipid bilayer. The results of calculations were verified against experimental studies of spatial arrangement of transmembrane and peripheral proteins in membranes.

The International Society for Computational Biology (ISCB) is a scholarly society for researchers in computational biology and bioinformatics. The society was founded in 1997 to provide a stable financial home for the Intelligent Systems for Molecular Biology (ISMB) conference and has grown to become a larger society working towards advancing understanding of living systems through computation and for communicating scientific advances worldwide.
Protein function prediction methods are techniques that bioinformatics researchers use to assign biological or biochemical roles to proteins. These proteins are usually ones that are poorly studied or predicted based on genomic sequence data. These predictions are often driven by data-intensive computational procedures. Information may come from nucleic acid sequence homology, gene expression profiles, protein domain structures, text mining of publications, phylogenetic profiles, phenotypic profiles, and protein-protein interaction. Protein function is a broad term: the roles of proteins range from catalysis of biochemical reactions to transport to signal transduction, and a single protein may play a role in multiple processes or cellular pathways.
The ISCB Overton Prize is a computational biology prize awarded annually for outstanding accomplishment by a scientist in the early to mid stage of his or her career. Laureates have made significant contribution to the field of computational biology either through research, education, service, or a combination of the three.
The ISCB Accomplishment by a Senior Scientist Award is an annual prize awarded by the International Society for Computational Biology for contributions to the field of computational biology.

David Tudor Jones is a Professor of Bioinformatics, and Head of Bioinformatics Group in the University College London. He is also the director in Bloomsbury Center for Bioinformatics, which is a joint Research Centre between UCL and Birkbeck, University of London and which also provides bioinformatics training and support services to biomedical researchers. In 2013, he is a member of editorial boards for PLoS ONE, BioData Mining, Advanced Bioinformatics, Chemical Biology & Drug Design, and Protein: Structure, Function and Bioinformatics.

The European Conference on Computational Biology (ECCB) is a scientific meeting on the subjects of bioinformatics and computational biology. It covers a wide spectrum of disciplines, including bioinformatics, computational biology, genomics, computational structural biology, and systems biology. ECCB is organized annually in different European cities. Since 2007, the conference has been held jointly with Intelligent Systems for Molecular Biology (ISMB) every second year. The conference also hosts the European ISCB Student Council Symposium. The proceedings of the conference are published by the journal Bioinformatics.

Chris Sander is a computational biologist based at the Dana-Farber Cancer Center and Harvard Medical School. Previously he was chair of the Computational Biology Programme at the Memorial Sloan–Kettering Cancer Center in New York City. In 2015, he moved his lab to the Dana–Farber Cancer Institute and the Cell Biology Department at Harvard Medical School.

Alfonso Valencia is a Spanish biologist, ICREA Professor, current director of the Life Sciences department at Barcelona Supercomputing Center. and of Spanish National Bioinformatics Institute (INB-ISCIII). From 2015-2018, he was President of the International Society for Computational Biology. His research is focused on the study of biomedical systems with computational biology and bioinformatics approaches.

Debora S. Marks is a researcher in computational biology and a Professor of Systems Biology at Harvard Medical School. Her research uses computational approaches to address a variety of biological problems.
Mona Singh is the Wang Family Professor in Computer Science in the Lewis-Sigler Institute for Integrative Genomics and the Department of Computer Science at Princeton University. She is Editor-in-Chief of the Journal of Computational Biology.

Hanah Margalit is a Professor in the faculty of medicine at the Hebrew University of Jerusalem. Her research combines bioinformatics, computational biology and systems biology, specifically in the fields of gene regulation in bacteria and eukaryotes.
Rita Casadio is an Adjunct Professor of Biochemistry/Biophysics in the Department of Pharmacy and Biotechnology at the University of Bologna.
Zhiping Weng is the Li Weibo Professor of biomedical research and chair of the program in integrative biology and bioinformatics at the University of Massachusetts Medical School. She was awarded Fellowship of the International Society for Computational Biology (ISCB) in 2020 for outstanding contributions to computational biology and bioinformatics.











