Mitochondrially encoded 12S ribosomal RNA is the SSU rRNA of the mitochondrial ribosome. In humans, 12S is encoded by the MT-RNR1 gene and is 959 nucleotides long. MT-RNR1 is one of the 37 genes contained in animal mitochondria genomes. Their 2 rRNA, 22 tRNA and 13 mRNA genes are very useful in phylogenetic studies, in particular the 12S and 16S rRNAs. The 12S rRNA is the mitochondrial homologue of the prokaryotic 16S and eukaryotic nuclear 18S ribosomal RNAs. Mutations in the MT-RNR1 gene may be associated with hearing loss. The rRNA gene also encodes a peptide MOTS-c, also known as Mitochondrial-derived peptide MOTS-c or Mitochondrial open reading frame of the 12S rRNA-c.

MT-ND6 is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 6 protein (ND6). The ND6 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variations in the human MT-ND6 gene are associated with Leigh's syndrome, Leber's hereditary optic neuropathy (LHON) and dystonia.

MT-ND4 is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 4 (ND4) protein. The ND4 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variations in the MT-ND4 gene are associated with age-related macular degeneration (AMD), Leber's hereditary optic neuropathy (LHON), mesial temporal lobe epilepsy (MTLE) and cystic fibrosis.

MT-ND2 is a gene of the mitochondrial genome coding for the NADH dehydrogenase 2 (ND2) protein. The ND2 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variants of human MT-ND2 are associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS), Leigh's syndrome (LS), Leber's hereditary optic neuropathy (LHON) and increases in adult BMI.

MT-ND4L is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 4L (ND4L) protein. The ND4L protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variants of human MT-ND4L are associated with increased BMI in adults and Leber's Hereditary Optic Neuropathy (LHON).

MT-ATP8 is a mitochondrial gene with the full name 'mitochondrially encoded ATP synthase membrane subunit 8' that encodes a subunit of mitochondrial ATP synthase, ATP synthase Fo subunit 8. This subunit belongs to the Fo complex of the large, transmembrane F-type ATP synthase. This enzyme, which is also known as complex V, is responsible for the final step of oxidative phosphorylation in the electron transport chain. Specifically, one segment of ATP synthase allows positively charged ions, called protons, to flow across a specialized membrane inside mitochondria. Another segment of the enzyme uses the energy created by this proton flow to convert a molecule called adenosine diphosphate (ADP) to ATP. Subunit 8 differs in sequence between Metazoa, plants and Fungi.

MT-ND1 is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 1 (ND1) protein. The ND1 protein is a subunit of NADH dehydrogenase, which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variants of the human MT-ND1 gene are associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS), Leigh's syndrome (LS), Leber's hereditary optic neuropathy (LHON) and increases in adult BMI.

Cytochrome c oxidase I (COX1) also known as mitochondrially encoded cytochrome c oxidase I (MT-CO1) is a protein that is encoded by the MT-CO1 gene in eukaryotes. The gene is also called COX1, CO1, or COI. Cytochrome c oxidase I is the main subunit of the cytochrome c oxidase complex. In humans, mutations in MT-CO1 have been associated with Leber's hereditary optic neuropathy (LHON), acquired idiopathic sideroblastic anemia, Complex IV deficiency, colorectal cancer, sensorineural deafness, and recurrent myoglobinuria.

Cytochrome c oxidase subunit III (COX3) is an enzyme that in humans is encoded by the MT-CO3 gene. It is one of main transmembrane subunits of cytochrome c oxidase. It is also one of the three mitochondrial DNA (mtDNA) encoded subunits of respiratory complex IV. Variants of it have been associated with isolated myopathy, severe encephalomyopathy, Leber hereditary optic neuropathy, mitochondrial complex IV deficiency, and recurrent myoglobinuria.

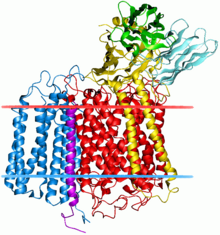
Cytochrome b is a protein that in humans is encoded by the MT-CYB gene. Its gene product is a subunit of the respiratory chain protein ubiquinol–cytochrome c reductase, which consists of the products of one mitochondrially encoded gene, MT-CYB, and ten nuclear genes—UQCRC1, UQCRC2, CYC1, UQCRFS1, UQCRB, "11kDa protein", UQCRH, Rieske protein presequence, "cyt c1 associated protein", and Rieske-associated protein.

SCO2 cytochrome c oxidase assembly is a protein that in humans is encoded by the SCO2 gene. The encoded protein is one of the cytochrome c oxidase (COX)(Complex IV) assembly factors. Human COX is a multimeric protein complex that requires several assembly factors. Cytochrome c oxidase (COX) catalyzes the transfer of electrons from cytochrome c to molecular oxygen, which helps to maintain the proton gradient across the inner mitochondrial membrane that is necessary for aerobic ATP production. The encoded protein is a metallochaperone that is involved in the biogenesis of cytochrome c oxidase subunit II. Mutations in this gene are associated with fatal infantile encephalocardiomyopathy and myopia 6.

Protein SCO1 homolog, mitochondrial, also known as SCO1, cytochrome c oxidase assembly protein, is a protein that in humans is encoded by the SCO1 gene. SCO1 localizes predominantly to blood vessels, whereas SCO2 is barely detectable, as well as to tissues with high levels of oxidative phosphorylation. The expression of SCO2 is also much higher than that of SCO1 in muscle tissue, while SCO1 is expressed at higher levels in liver tissue than SCO2. Mutations in both SCO1 and SCO2 are associated with distinct clinical phenotypes as well as tissue-specific cytochrome c oxidase deficiency.

Cytochrome c oxidase subunit 4 isoform 1, mitochondrial (COX4I1) is an enzyme that in humans is encoded by the COX4I1 gene. COX4I1 is a nuclear-encoded isoform of cytochrome c oxidase (COX) subunit 4. Cytochrome c oxidase is a multi-subunit enzyme complex that couples the transfer of electrons from cytochrome c to molecular oxygen and contributes to a proton electrochemical gradient across the inner mitochondrial membrane, acting as the terminal enzyme of the mitochondrial respiratory chain. Antibodies against COX4 can be used to identify the inner membrane of mitochondria in immunofluorescence studies. Mutations in COX4I1 have been associated with COX deficiency and Fanconi anemia.

Cytochrome c oxidase subunit 6B1 is an enzyme that in humans is encoded by the COX6B1 gene. Cytochrome c oxidase 6B1 is a subunit of the cytochrome c oxidase complex, also known as Complex IV, the last enzyme in the mitochondrial electron transport chain. Mutations of the COX6B1 gene are associated with severe infantile encephalomyopathy and mitochondrial complex IV deficiency (MT-C4D).

Cytochrome c oxidase assembly protein COX15 homolog (COX15), also known as heme A synthase, is a protein that in humans is encoded by the COX15 gene. This protein localizes to the inner mitochondrial membrane and involved in heme A biosynthesis. COX15 is also part of a three-component mono-oxygenase that catalyses the hydroxylation of the methyl group at position eight of the protoheme molecule. Mutations in this gene has been reported in patients with hypertrophic cardiomyopathy as well as Leigh syndrome, and characterized by delayed onset of symptoms, hypotonia, feeding difficulties, failure to thrive, motor regression, and brain stem signs.
Mitochondrially encoded tRNA aspartic acid also known as MT-TD is a transfer RNA which in humans is encoded by the mitochondrial MT-TD gene.
Mitochondrially encoded tRNA phenylalanine also known as MT-TF is a transfer RNA which in humans is encoded by the mitochondrial MT-TF gene.
Mitochondrially encoded tRNA leucine 2 (CUN) also known as MT-TL2 is a transfer RNA which in humans is encoded by the mitochondrial MT-TL2 gene.
Mitochondrially encoded tRNA asparagine also known as MT-TN is a transfer RNA which in humans is encoded by the mitochondrial MT-TN gene.
Mitochondrially encoded tRNA arginine also known as MT-TR is a transfer RNA which in humans is encoded by the mitochondrial MT-TR gene.