
In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

Lipoxygenases are a family of (non-heme) iron-containing enzymes most of which catalyze the dioxygenation of polyunsaturated fatty acids in lipids containing a cis,cis-1,4- pentadiene into cell signaling agents that serve diverse roles as autocrine signals that regulate the function of their parent cells, paracrine signals that regulate the function of nearby cells, and endocrine signals that regulate the function of distant cells.
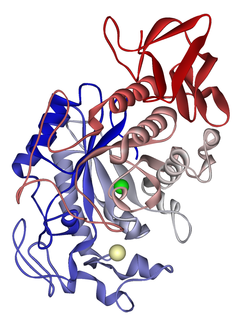
Alpha-amylase(α-amylase) is an enzyme that hydrolyses alpha bonds of large, alpha-linked polysaccharides, such as starch and glycogen, yielding shorter chains thereof, dextrins, and maltose. It is the major form of amylase found in humans and other mammals. It is also present in seeds containing starch as a food reserve, and is secreted by many fungi. It is a member of glycoside hydrolase family 13.

In enzymology, an alanine racemase is an enzyme that catalyzes the chemical reaction

A cutinase (EC 3.1.1.74) is an enzyme that catalyzes the chemical reaction

Proteins that bind cyclic nucleotides share a structural domain of about 120 residues. The best studied of these proteins is the prokaryotic catabolite gene activator where such a domain is known to be composed of three alpha-helices and a distinctive eight-stranded, antiparallel beta-barrel structure. There are six invariant amino acids in this domain, three of which are glycine residues that are thought to be essential for maintenance of the structural integrity of the beta-barrel. cAMP- and cGMP-dependent protein kinases contain two tandem copies of the cyclic nucleotide-binding domain. The cAPK's are composed of two different subunits, a catalytic chain and a regulatory chain, which contains both copies of the domain. The cGPK's are single chain enzymes that include the two copies of the domain in their N-terminal section. Vertebrate cyclic nucleotide-gated ion-channels also contain this domain. Two such cations channels have been fully characterized, one is found in rod cells where it plays a role in visual signal transduction.
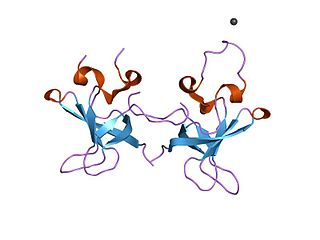
The S1 domain is a protein domain that was originally identified in ribosomal protein S1 but is found in a large number of RNA-associated proteins. The structure of the S1 RNA-binding domain from the Escherichia coli polynucleotide phosphorylase has been determined using NMR methods and consists of a five-stranded antiparallel beta barrel. Conserved residues on one face of the barrel and adjacent loops form the putative RNA-binding site.

In molecular biology, the ACT domain is a protein domain that is found in a variety of proteins involved in metabolism. ACT domains are linked to a wide range of metabolic enzymes that are regulated by amino acid concentration. The ACT domain is named after three of the proteins that contain it: aspartate kinase, chorismate mutase and TyrA. The archetypical ACT domain is the C-terminal regulatory domain of 3-phosphoglycerate dehydrogenase (3PGDH), which folds with a ferredoxin-like topology. A pair of ACT domains form an eight-stranded antiparallel sheet with two molecules of allosteric inhibitor serine bound in the interface. Biochemical exploration of a few other proteins containing ACT domains supports the suggestions that these domains contain the archetypical ACT structure.

In molecular biology, the AMMECR1 protein is a protein encoded by the AMMECR1 gene on human chromosome Xq22.3.
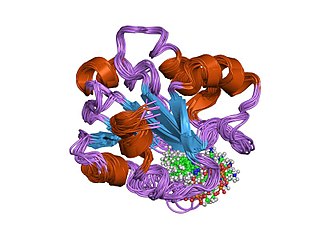
In molecular biology, the vitamin B12-binding domain is a protein domain which binds to cobalamin. It can bind two different forms of the cobalamin cofactor, with cobalt bonded either to a methyl group (methylcobalamin) or to 5'-deoxyadenosine (adenosylcobalamin). Cobalamin-binding domains are mainly found in two families of enzymes present in animals and prokaryotes, which perform distinct kinds of reactions at the cobalt-carbon bond. Enzymes that require methylcobalamin carry out methyl transfer reactions. Enzymes that require adenosylcobalamin catalyse reactions in which the first step is the cleavage of adenosylcobalamin to form cob(II)alamin and the 5'-deoxyadenosyl radical, and thus act as radical generators. In both types of enzymes the B12-binding domain uses a histidine to bind the cobalt atom of cobalamin cofactors. This histidine is embedded in a DXHXXG sequence, the most conserved primary sequence motif of the domain. Proteins containing the cobalamin-binding domain include:

In molecular biology, a carbohydrate-binding module (CBM) is a protein domain found in carbohydrate-active enzymes. The majority of these domains have carbohydrate-binding activity. Some of these domains are found on cellulosomal scaffoldin proteins. CBMs were previously known as cellulose-binding domains. CBMs are classified into numerous families, based on amino acid sequence similarity. There are currently 64 families of CBM in the CAZy database.

Ferredoxin-thioredoxin reductase EC 1.8.7.2, systematic name ferredoxin:thioredoxin disulfide oxidoreductase, is a [4Fe-4S] protein that plays an important role in the ferredoxin/thioredoxin regulatory chain. It catalyzes the following reaction:

In molecular biology, Glycoside hydrolase family 14 is a family of glycoside hydrolases.

In molecular biology, Glycoside hydrolase family 2 is a family of glycoside hydrolases.

In molecular biology, glycoside hydrolase family 42 is a family of glycoside hydrolases.

In molecular biology, glycoside hydrolase family 65 is a family of glycoside hydrolases.
In molecular biology, glycoside hydrolase family 97 is a family of glycoside hydrolases.

In molecular biology, group III pyridoxal-dependent decarboxylases are a family of bacterial enzymes comprising ornithine decarboxylase EC 4.1.1.17, lysine decarboxylase EC 4.1.1.18 and arginine decarboxylase EC 4.1.1.19.

In molecular biology, the protein domain, Ydc2, is a Holliday junction resolvase from the fission yeast Schizosaccharomyces pombe that is involved in the maintenance of mitochondrial DNA.



















