
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA, which then may undergo error-prone repair, cause an error during other forms of repair, or cause an error during replication. Mutations may also result from substitution, insertion or deletion of segments of DNA due to mobile genetic elements.

A transposable element (TE), also transposon, or jumping gene, is a type of mobile genetic element, a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size.

Genetic recombination is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryotes, genetic recombination during meiosis can lead to a novel set of genetic information that can be further passed on from parents to offspring. Most recombination occurs naturally and can be classified into two types: (1) interchromosomal recombination, occurring through independent assortment of alleles whose loci are on different but homologous chromosomes ; & (2) intrachromosomal recombination, occurring through crossing over.
Molecular evolution describes how inherited DNA and/or RNA change over evolutionary time, and the consequences of this for proteins and other components of cells and organisms. Molecular evolution is the basis of phylogenetic approaches to describing the tree of life. Molecular evolution overlaps with population genetics, especially on shorter timescales. Topics in molecular evolution include the origins of new genes, the genetic nature of complex traits, the genetic basis of adaptation and speciation, the evolution of development, and patterns and processes underlying genomic changes during evolution.

Pseudogenes are nonfunctional segments of DNA that resemble functional genes. Pseudogenes can be formed from both protein-coding genes and non-coding genes. In the case of protein-coding genes, most pseudogenes arise as superfluous copies of functional genes, either directly by gene duplication or indirectly by reverse transcription of an mRNA transcript. Pseudogenes are usually identified when genome sequence analysis finds gene-like sequences that lack regulatory sequences or are incapable of producing a functional product. Pseudogenes are a type of junk DNA.
Viral evolution is a subfield of evolutionary biology and virology that is specifically concerned with the evolution of viruses. Viruses have short generation times, and many—in particular RNA viruses—have relatively high mutation rates. Although most viral mutations confer no benefit and often even prove deleterious to viruses, the rapid rate of viral mutation combined with natural selection allows viruses to quickly adapt to changes in their host environment. In addition, because viruses typically produce many copies in an infected host, mutated genes can be passed on to many offspring quickly. Although the chance of mutations and evolution can change depending on the type of virus, viruses overall have high chances for mutations.

A gene family is a set of several similar genes, formed by duplication of a single original gene, and generally with similar biochemical functions. One such family are the genes for human hemoglobin subunits; the ten genes are in two clusters on different chromosomes, called the α-globin and β-globin loci. These two gene clusters are thought to have arisen as a result of a precursor gene being duplicated approximately 500 million years ago.
The ribosomal DNA consists of a group of ribosomal RNA encoding genes and related regulatory elements, and is widespread in similar configuration in all domains of life. The ribosomal DNA encodes the non-coding ribosomal RNA, integral structural elements in the assembly of ribosomes, its importance making it the most abundant section of RNA found in cells of eukaryotes. Additionally, these segments includes regulatory sections, such as a promotor specific to the RNA polymerase I, as well as both transcribed and non-transcribed spacer segments.
Gene conversion is the process by which one DNA sequence replaces a homologous sequence such that the sequences become identical after the conversion. Gene conversion can be either allelic, meaning that one allele of the same gene replaces another allele, or ectopic, meaning that one paralogous DNA sequence converts another.

A nuclear gene is a gene that has its DNA nucleotide sequence physically situated within the cell nucleus of a eukaryotic organism. This term is employed to differentiate nuclear genes, which are located in the cell nucleus, from genes that are found in mitochondria or chloroplasts. The vast majority of genes in eukaryotes are nuclear.

In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes. During gene expression, DNA is first copied into RNA. RNA can be directly functional or be the intermediate template for the synthesis of a protein.
Gabriel A. Dover was a British geneticist, best known for coining the term molecular drive in 1982 to describe a putative third evolutionary force operating distinctly from natural selection and genetic drift.
Exon shuffling is a molecular mechanism for the formation of new genes. It is a process through which two or more exons from different genes can be brought together ectopically, or the same exon can be duplicated, to create a new exon-intron structure. There are different mechanisms through which exon shuffling occurs: transposon mediated exon shuffling, crossover during sexual recombination of parental genomes and illegitimate recombination.

Masatoshi Nei was a Japanese-born American evolutionary biologist.

A gene cluster is a group of two or more genes found within an organism's DNA that encode similar polypeptides or proteins which collectively share a generalized function and are often located within a few thousand base pairs of each other. The size of gene clusters can vary significantly, from a few genes to several hundred genes. Portions of the DNA sequence of each gene within a gene cluster are found to be identical; however, the protein encoded by each gene is distinct from the proteins encoded by the other genes within the cluster. Gene clusters often result from expansions of a single gene caused by repeated duplication events, and may be observed near one another on the same chromosome or on different, but homologous chromosomes. An example of a gene cluster is the Hox gene, which is made up of eight genes and is part of the Homeobox gene family.
18S ribosomal RNA is a part of the ribosomal RNA in eukaryotes. It is a component of the Eukaryotic small ribosomal subunit (40S) and the cytosolic homologue of both the 12S rRNA in mitochondria and the 16S rRNA in plastids and prokaryotes. Similar to the prokaryotic 16S rRNA, the genes of the 18S ribosomal RNA have been widely used for phylogenetic studies and biodiversity screening of eukaryotes.

Gene redundancy is the existence of multiple genes in the genome of an organism that perform the same function. Gene redundancy can result from gene duplication. Such duplication events are responsible for many sets of paralogous genes. When an individual gene in such a set is disrupted by mutation or targeted knockout, there can be little effect on phenotype as a result of gene redundancy, whereas the effect is large for the knockout of a gene with only one copy. Gene knockout is a method utilized in some studies aiming to characterize the maintenance and fitness effects functional overlap.
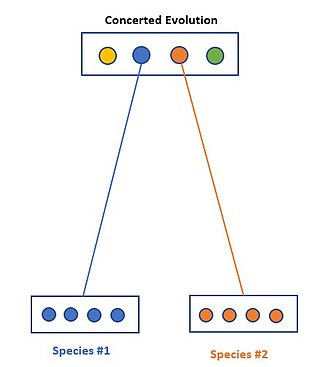
Concerted evolution is the phenomenon where paralogous genes within one species are more closely related to one another than to members of the same gene family in closely related species. In other terms, when specific members of a family are investigated, a greater amount of similarity is found within a species rather than between species. This is suggesting that members within this family do not in fact evolve independently of one another.
Evolution of cells refers to the evolutionary origin and subsequent evolutionary development of cells. Cells first emerged at least 3.8 billion years ago approximately 750 million years after Earth was formed.

Genome evolution is the process by which a genome changes in structure (sequence) or size over time. The study of genome evolution involves multiple fields such as structural analysis of the genome, the study of genomic parasites, gene and ancient genome duplications, polyploidy, and comparative genomics. Genome evolution is a constantly changing and evolving field due to the steadily growing number of sequenced genomes, both prokaryotic and eukaryotic, available to the scientific community and the public at large.











