
Chlorophyll is any of several related green pigments found in cyanobacteria and in the chloroplasts of algae and plants. Its name is derived from the Greek words χλωρός, khloros and φύλλον, phyllon ("leaf"). Chlorophyll allow plants to absorb energy from light.

Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other, nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD+ and NADH (H for hydrogen), respectively.
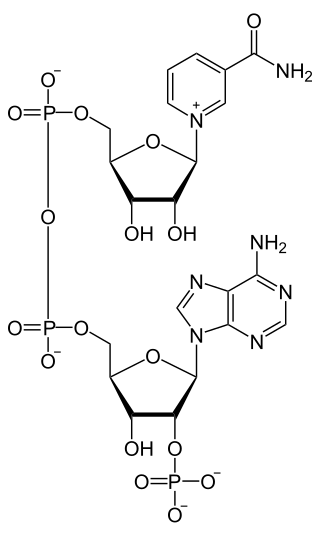
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP+ or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). NADPH is the reduced form of NADP+, the oxidized form. NADP+ is used by all forms of cellular life.
Nitrogenases are enzymes (EC 1.18.6.1EC 1.19.6.1) that are produced by certain bacteria, such as cyanobacteria (blue-green bacteria) and rhizobacteria. These enzymes are responsible for the reduction of nitrogen (N2) to ammonia (NH3). Nitrogenases are the only family of enzymes known to catalyze this reaction, which is a step in the process of nitrogen fixation. Nitrogen fixation is required for all forms of life, with nitrogen being essential for the biosynthesis of molecules (nucleotides, amino acids) that create plants, animals and other organisms. They are encoded by the Nif genes or homologs. They are related to protochlorophyllide reductase.

Photosystem I is one of two photosystems in the photosynthetic light reactions of algae, plants, and cyanobacteria. Photosystem I is an integral membrane protein complex that uses light energy to catalyze the transfer of electrons across the thylakoid membrane from plastocyanin to ferredoxin. Ultimately, the electrons that are transferred by Photosystem I are used to produce the moderate-energy hydrogen carrier NADPH. The photon energy absorbed by Photosystem I also produces a proton-motive force that is used to generate ATP. PSI is composed of more than 110 cofactors, significantly more than Photosystem II.
Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.
Iron–sulfur proteins are proteins characterized by the presence of iron–sulfur clusters containing sulfide-linked di-, tri-, and tetrairon centers in variable oxidation states. Iron–sulfur clusters are found in a variety of metalloproteins, such as the ferredoxins, as well as NADH dehydrogenase, hydrogenases, coenzyme Q – cytochrome c reductase, succinate – coenzyme Q reductase and nitrogenase. Iron–sulfur clusters are best known for their role in the oxidation-reduction reactions of electron transport in mitochondria and chloroplasts. Both Complex I and Complex II of oxidative phosphorylation have multiple Fe–S clusters. They have many other functions including catalysis as illustrated by aconitase, generation of radicals as illustrated by SAM-dependent enzymes, and as sulfur donors in the biosynthesis of lipoic acid and biotin. Additionally, some Fe–S proteins regulate gene expression. Fe–S proteins are vulnerable to attack by biogenic nitric oxide, forming dinitrosyl iron complexes. In most Fe–S proteins, the terminal ligands on Fe are thiolate, but exceptions exist.

In enzymology, a shikimate dehydrogenase (EC 1.1.1.25) is an enzyme that catalyzes the chemical reaction

In enzymology, divinyl chlorophyllide a 8-vinyl-reductase (EC 1.3.1.75) is an enzyme that catalyzes the chemical reaction
In enzymology, a phycocyanobilin:ferredoxin oxidoreductase is an enzyme that catalyzes the chemical reaction
In enzymology, a phytochromobilin:ferredoxin oxidoreductase is an enzyme that catalyzes the chemical reaction
In enzymology, a ferredoxin-NADP+ reductase (EC 1.18.1.2) abbreviated FNR, is an enzyme that catalyzes the chemical reaction
In enzymology, a dephospho-[reductase kinase] kinase is an enzyme that catalyzes the chemical reaction

Protochlorophyllide, or monovinyl protochlorophyllide, is an intermediate in the biosynthesis of chlorophyll a. It lacks the phytol side-chain of chlorophyll and the reduced pyrrole in ring D. Protochlorophyllide is highly fluorescent; mutants that accumulate it glow red if irradiated with blue light. In angiosperms, the later steps which convert protochlorophyllide to chlorophyll are light-dependent, and such plants are pale (chlorotic) if grown in the darkness. Gymnosperms, algae, and photosynthetic bacteria have another, light-independent enzyme and grow green in the darkness as well.

Light-dependent reactions is jargon for certain photochemical reactions that are involved in photosynthesis, the main process by which plants acquire energy. There are two light dependent reactions, the first occurs at photosystem II (PSII) and the second occurs at photosystem I (PSI),
Chlorophyll(ide) b reductase (EC 1.1.1.294), chlorophyll b reductase, Chl b reductase) is an enzyme with systematic name 71-hydroxychlorophyllide-a:NAD(P)+ oxidoreductase. This enzyme catalyses the following chemical reaction

Chlorophyllide-a oxygenase (EC 1.14.13.122), chlorophyllide a oxygenase, chlorophyll-b synthase, CAO) is an enzyme with systematic name chlorophyllide-a:oxygen 7-oxidoreductase. This enzyme catalyses the following chemical reactions

Magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase, is an enzyme with systematic name magnesium-protoporphyrin-IX 13-monomethyl ester, ferredoxin:oxygen oxidoreductase (hydroxylating). In plants this enzyme catalyses the following overall chemical reaction

Chlorophyllide a and Chlorophyllide b are the biosynthetic precursors of chlorophyll a and chlorophyll b respectively. Their propionic acid groups are converted to phytyl esters by the enzyme chlorophyll synthase in the final step of the pathway. Thus the main interest in these chemical compounds has been in the study of chlorophyll biosynthesis in plants, algae and cyanobacteria. Chlorophyllide a is also an intermediate in the biosynthesis of bacteriochlorophylls.

Chlorophyllide a reductase (EC 1.3.7.15), also known as COR, is an enzyme with systematic name bacteriochlorophyllide-a:ferredoxin 7,8-oxidoreductase. It catalyses the following chemical reaction













