Related Research Articles

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.

DD-transpeptidase is a bacterial enzyme that catalyzes the transfer of the R-L-αα-D-alanyl moiety of R-L-αα-D-alanyl-D-alanine carbonyl donors to the γ-OH of their active-site serine and from this to a final acceptor. It is involved in bacterial cell wall biosynthesis, namely, the transpeptidation that crosslinks the peptide side chains of peptidoglycan strands.
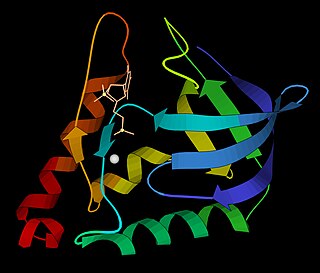
Micrococcal nuclease is an endo-exonuclease that preferentially digests single-stranded nucleic acids. The rate of cleavage is 30 times greater at the 5' side of A or T than at G or C and results in the production of mononucleotides and oligonucleotides with terminal 3'-phosphates. The enzyme is also active against double-stranded DNA and RNA and all sequences will be ultimately cleaved.
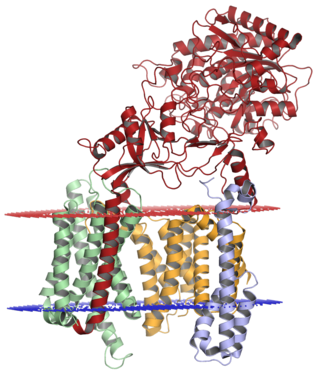
Gamma secretase is a multi-subunit protease complex, itself an integral membrane protein, that cleaves single-pass transmembrane proteins at residues within the transmembrane domain. Proteases of this type are known as intramembrane proteases. The most well-known substrate of gamma secretase is amyloid precursor protein, a large integral membrane protein that, when cleaved by both gamma and beta secretase, produces a short 37-43 amino acid peptide called amyloid beta whose abnormally folded fibrillar form is the primary component of amyloid plaques found in the brains of Alzheimer's disease patients. Gamma secretase is also critical in the related processing of several other type I integral membrane proteins, such as Notch, ErbB4, E-cadherin, N-cadherin, ephrin-B2, or CD44.

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.
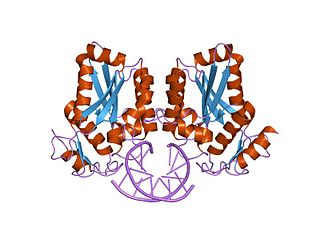
BamHI is a type II restriction endonuclease, having the capacity for recognizing short sequences of DNA and specifically cleaving them at a target site. This exhibit focuses on the structure-function relations of BamHI as described by Newman, et al. (1995). BamHI binds at the recognition sequence 5'-GGATCC-3', and cleaves these sequences just after the 5'-guanine on each strand. This cleavage results in sticky ends which are 4 bp long. In its unbound form, BamHI displays a central b sheet, which resides in between α-helices.
Pantothenate kinase (EC 2.7.1.33, PanK; CoaA) is the first enzyme in the Coenzyme A (CoA) biosynthetic pathway. It phosphorylates pantothenate (vitamin B5) to form 4'-phosphopantothenate at the expense of a molecule of adenosine triphosphate (ATP). It is the rate-limiting step in the biosynthesis of CoA.
Toxic shock syndrome toxin-1 (TSST-1) is a superantigen with a size of 22 kDa produced by 5 to 25% of Staphylococcus aureus isolates. It causes toxic shock syndrome (TSS) by stimulating the release of large amounts of interleukin-1, interleukin-2 and tumour necrosis factor. In general, the toxin is not produced by bacteria growing in the blood; rather, it is produced at the local site of an infection, and then enters the blood stream.

A Disintegrin and metalloproteinase domain-containing protein 10, also known as ADAM10 or CDw156 or CD156c is a protein that in humans is encoded by the ADAM10 gene.

Phospholipase C (PLC) is a class of membrane-associated enzymes that cleave phospholipids just before the phosphate group (see figure). It is most commonly taken to be synonymous with the human forms of this enzyme, which play an important role in eukaryotic cell physiology, in particular signal transduction pathways. Phospholipase C's role in signal transduction is its cleavage of phosphatidylinositol 4,5-bisphosphate (PIP2) into diacyl glycerol (DAG) and inositol 1,4,5-trisphosphate (IP3), which serve as second messengers. Activators of each PLC vary, but typically include heterotrimeric G protein subunits, protein tyrosine kinases, small G proteins, Ca2+, and phospholipids.

Methionine-R-sulfoxide reductase B1 is an enzyme that in humans is encoded by the SEPX1 gene.
Fibronectin binding protein A (FnBPA) is a Staphylococcus aureus MSCRAMM cell surface-bound protein that binds to both fibronectin and fibrinogen.

Dispersin B is a 40 kDa glycoside hydrolase produced by the periodontal pathogen, Aggregatibacter actinomycetemcomitans. The bacteria secrete Dispersin B to release adherent cells from a mature biofilm colony by disrupting biofilm formation. The enzyme catalyzes the hydrolysis of linear polymers of N-acetyl-D-glucosamines found in the biofilm matrices. Poly-acetyl glucosamines are integral to the structural integrity of the biofilms of various Gram-positive bacteria and Gram-negative bacteria and are referred to as PIA (PNAG,PS/A) in Staphylococcus species and PGA in Escherichia coli. By degrading the biofilm matrix, Dispersin B allows for the release of bacterial cells that can adhere to new surfaces close by and extend the biofilm or start new colonies. Currently there is interest in Dispersin B as a commercial anti-biofilm agent that could be combined with antibiotics for the treatment of bacterial infections.
Sortase refers to a group of prokaryotic enzymes that modify surface proteins by recognizing and cleaving a carboxyl-terminal sorting signal. For most substrates of sortase enzymes, the recognition signal consists of the motif LPXTG (Leu-Pro-any-Thr-Gly), then a highly hydrophobic transmembrane sequence, followed by a cluster of basic residues such as arginine. Cleavage occurs between the Thr and Gly, with transient attachment through the Thr residue to the active site Cys residue, followed by transpeptidation that attaches the protein covalently to cell wall components. Sortases occur in almost all Gram-positive bacteria and the occasional Gram-negative bacterium or Archaea, where cell wall LPXTG-mediated decoration has not been reported. Although sortase A, the "housekeeping" sortase, typically acts on many protein targets, other forms of sortase recognize variant forms of the cleavage motif, or catalyze the assembly of pilins into pili.

Glutamyl endopeptidase is an extracellular bacterial serine protease of the glutamyl endopeptidase I family that was initially isolated from the Staphylococcus aureus strain V8. The protease is, hence, commonly referred to as "V8 protease", or alternatively SspA from its corresponding gene.
Sortases are membrane anchored enzyme that sort these surface proteins onto the bacterial cell surface and anchor them to the peptidoglycan. There are different types of sortases and each catalyse the anchoring of different proteins to cell walls.
LPXTGase refers to an endopeptidase enzyme from Streptococci and Staphylococci with the capacity to cleave the carboxy-terminal LPXTG anchor motif of surface proteins similar to Sortase. However, LPXTGase differs significantly from Sortase in several ways: a) it is glycosylated, b) it contains unconventional amino acids, and c) it contains D-amino acids. The latter two characteristics indicate that ribosomes are not involve in the synthesis of LPXTGase. Data suggest that the enzymes responsible for cell wall assembly also assemble LPXTGase.
Antivirulence is the concept of blocking virulence factors. In regards to bacteria, the idea is to design agents that block virulence rather than kill bacteria en masse, as the current regime results in much more selective pressure.
A protein-sorting transpeptidase is an enzyme, such as the sortase SrtA of Staphylococcus aureus, that cleaves one or more target proteins produced by the same cell, as part of a specialized pathway of protein targeting. The typical prokaryotic protein-sorting transpeptidase is characterized as a protease, but does not simply hydrolyze a peptide bond. Instead, the larger, N-terminal portion of the cleaved polypeptide is transferred onto another molecule, such as a precursor of the peptidoglycan cell wall in Gram-positive bacteria.
Olaf Schneewind was a German-born American microbiologist who made important contributions to the study of bacterial cell wall composition and assembly as well as the pathogenesis of the microbial species S. aureus. He was elected to the National Academy of Sciences in 2018.
References
- ↑ Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O (October 1999). "Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif". Proceedings of the National Academy of Sciences of the United States of America. 96 (22): 12424–9. Bibcode:1999PNAS...9612424T. doi: 10.1073/pnas.96.22.12424 . PMC 22937 . PMID 10535938.
- ↑ Zong Y, Bice TW, Ton-That H, Schneewind O, Narayana SV (July 2004). "Crystal structures of Staphylococcus aureus sortase A and its substrate complex". The Journal of Biological Chemistry. 279 (30): 31383–9. doi: 10.1074/jbc.m401374200 . PMID 15117963.
- ↑ Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD, Sessions RB, Kehoe MA, McCafferty DG, Banfield MJ (March 2009). "Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism". The Journal of Biological Chemistry. 284 (11): 6924–33. doi: 10.1074/jbc.m805406200 . PMC 2652338 . PMID 19129180.
- ↑ Suree N, Liew CK, Villareal VA, Thieu W, Fadeev EA, Clemens JJ, Jung ME, Clubb RT (September 2009). "The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized". The Journal of Biological Chemistry. 284 (36): 24465–77. doi: 10.1074/jbc.M109.022624 . PMC 2782039 . PMID 19592495.
- ↑ Popp MW, Antos JM, Ploegh HL (April 2009). "Site-specific protein labeling via sortase-mediated transpeptidation". Current Protocols in Protein Science. Chapter 15: 15.3.1–15.3.9. doi:10.1002/0471140864.ps1503s56. PMC 5551486 . PMID 19365788.
- ↑ Popp MW, Ploegh HL (May 2011). "Making and breaking peptide bonds: protein engineering using sortase". Angewandte Chemie. 50 (22): 5024–32. doi:10.1002/anie.201008267. PMID 21538739.