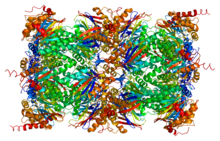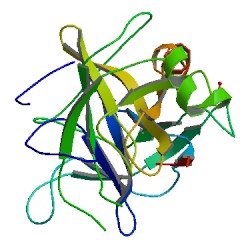
Chymotrypsin (EC 3.4.21.1, chymotrypsins A and B, alpha-chymar ophth, avazyme, chymar, chymotest, enzeon, quimar, quimotrase, alpha-chymar, alpha-chymotrypsin A, alpha-chymotrypsin) is a digestive enzyme component of pancreatic juice acting in the duodenum, where it performs proteolysis, the breakdown of proteins and polypeptides. Chymotrypsin preferentially cleaves peptide amide bonds where the side chain of the amino acid N-terminal to the scissile amide bond (the P1 position) is a large hydrophobic amino acid (tyrosine, tryptophan, and phenylalanine). These amino acids contain an aromatic ring in their side chain that fits into a hydrophobic pocket (the S1 position) of the enzyme. It is activated in the presence of trypsin. The hydrophobic and shape complementarity between the peptide substrate P1 side chain and the enzyme S1 binding cavity accounts for the substrate specificity of this enzyme. Chymotrypsin also hydrolyzes other amide bonds in peptides at slower rates, particularly those containing leucine at the P1 position.

Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion.

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in numerous biological pathways, including digestion of ingested proteins, protein catabolism, and cell signaling.

In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the binding site, and residues that catalyse a reaction of that substrate, the catalytic site. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes.

Serine proteases are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like.

A metalloproteinase, or metalloprotease, is any protease enzyme whose catalytic mechanism involves a metal. An example is ADAM12 which plays a significant role in the fusion of muscle cells during embryo development, in a process known as myogenesis.

DD-transpeptidase is a bacterial enzyme that catalyzes the transfer of the R-L-αα-D-alanyl moiety of R-L-αα-D-alanyl-D-alanine carbonyl donors to the γ-OH of their active-site serine and from this to a final acceptor. It is involved in bacterial cell wall biosynthesis, namely, the transpeptidation that crosslinks the peptide side chains of peptidoglycan strands.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.

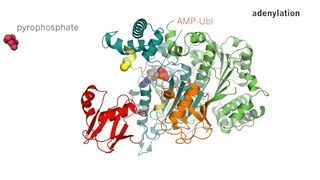
Deubiquitinating enzymes (DUBs), also known as deubiquitinating peptidases, deubiquitinating isopeptidases, deubiquitinases, ubiquitin proteases, ubiquitin hydrolases, or ubiquitin isopeptidases, are a large group of proteases that cleave ubiquitin from proteins. Ubiquitin is attached to proteins in order to regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions. DUBs can reverse these effects by cleaving the peptide or isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 100 DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases. The cysteine proteases comprise ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumour proteases (OTU). The metalloprotease group contains only the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain proteases.

Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad.
Enzyme catalysis is the increase in the rate of a process by a biological molecule, an "enzyme". Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, called the active site.
Serine hydrolases are one of the largest known enzyme classes comprising approximately ~200 enzymes or 1% of the genes in the human proteome. A defining characteristic of these enzymes is the presence of a particular serine at the active site, which is used for the hydrolysis of substrates. The hydrolysis of the ester or peptide bond proceeds in two steps. First, the acyl part of the substrate is transferred to the serine, making a new ester or amide bond and releasing the other part of the substrate is released. Later, in a slower step, the bond between the serine and the acyl group is hydrolyzed by water or hydroxide ion, regenerating free enzyme. Unlike other, non-catalytic, serines, the reactive serine of these hydrolases is typically activated by a proton relay involving a catalytic triad consisting of the serine, an acidic residue and a basic residue, although variations on this mechanism exist.

TEV protease is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV). It is a member of the PA clan of chymotrypsin-like proteases. Due to its high sequence specificity, TEV protease is frequently used for the controlled cleavage of fusion proteins in vitro and in vivo.
Carboxypeptidase A usually refers to the pancreatic exopeptidase that hydrolyzes peptide bonds of C-terminal residues with aromatic or aliphatic side-chains. Most scientists in the field now refer to this enzyme as CPA1, and to a related pancreatic carboxypeptidase as CPA2.

Carnitine O-acetyltransferase also called carnitine acetyltransferase is an enzyme that encoded by the CRAT gene that catalyzes the chemical reaction

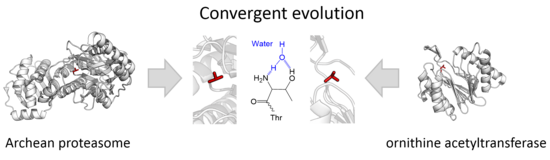
Subtilases are a family of subtilisin-like serine proteases. They appear to have independently and convergently evolved an Asp/Ser/His catalytic triad, like in the trypsin serine proteases. The structure of proteins in this family shows that they have an alpha/beta fold containing a 7-stranded parallel beta sheet.
A protein superfamily is the largest grouping (clade) of proteins for which common ancestry can be inferred. Usually this common ancestry is inferred from structural alignment and mechanistic similarity, even if no sequence similarity is evident. Sequence homology can then be deduced even if not apparent. Superfamilies typically contain several protein families which show sequence similarity within each family. The term protein clan is commonly used for protease and glycosyl hydrolases superfamilies based on the MEROPS and CAZy classification systems.

The PA clan is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have identity of <10%. The clan contains both cysteine and serine proteases. PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.

Glutamic proteases are a group of proteolytic enzymes containing a glutamic acid residue within the active site. This type of protease was first described in 2004 and became the sixth catalytic type of protease. Members of this group of protease had been previously assumed to be an aspartate protease, but structural determination showed it to belong to a novel protease family. The first structure of this group of protease was scytalidoglutamic peptidase, the active site of which contains a catalytic dyad, glutamic acid (E) and glutamine (Q), which give rise to the name eqolisin. This group of proteases are found primarily in pathogenic fungi affecting plant and human.
Asparagine peptide lyase are one of the seven groups in which proteases, also termed proteolytic enzymes, peptidases, or proteinases, are classified according to their catalytic residue. The catalytic mechanism of the asparagine peptide lyases involves an asparagine residue acting as nucleophile to perform a nucleophilic elimination reaction, rather than hydrolysis, to catalyse the breaking of a peptide bond.