Related Research Articles

Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases.

Ubiquitin is a small regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ubiquitously. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A.

Ubiquitin-like modifier activating enzyme 1 (UBA1) is an enzyme which in humans is encoded by the UBA1 gene. UBA1 participates in ubiquitination and the NEDD8 pathway for protein folding and degradation, among many other biological processes. This protein has been linked to X-linked spinal muscular atrophy type 2, neurodegenerative diseases, and cancers.

Deubiquitinating enzymes (DUBs), also known as deubiquitinating peptidases, deubiquitinating isopeptidases, deubiquitinases, ubiquitin proteases, ubiquitin hydrolases, ubiquitin isopeptidases, are a large group of proteases that cleave ubiquitin from proteins. Ubiquitin is attached to proteins in order to regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions. DUBs can reverse these effects by cleaving the peptide or isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 100 DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases. The cysteine proteases comprise ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumour proteases (OTU). The metalloprotease group contains only the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain proteases.
In molecular biology, SUMOproteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.
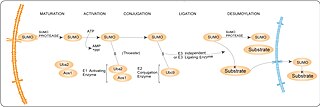
SUMO enzymatic cascade catalyzes the dynamic posttranslational modification process of sumoylation. The Small Ubiquitin-related Modifier, SUMO-1, is a ubiquitin-like family member that is conjugated to its substrates through three discrete enzymatic steps : activation, involving the E1 enzyme (SAE1/SAE2); conjugation, involving the E2 enzyme (UBE2I); substrate modification, through the cooperation of the E2 and E3 protein ligases.

Small ubiquitin-related modifier 1 is a protein that in humans is encoded by the SUMO1 gene.

SUMO-conjugating enzyme UBC9 is an enzyme that in humans is encoded by the UBE2I gene. It is also sometimes referred to as "ubiquitin conjugating enzyme E2I" or "ubiquitin carrier protein 9", even though these names do not accurately describe its function.

Small ubiquitin-related modifier 3 is a protein that in humans is encoded by the SUMO3 gene.

SUMO1/sentrin/SMT3 specific peptidase 3, also known as SENP3, is a protein which in humans is encoded by the SENP3 gene.

Sentrin-specific protease 1 is an enzyme that in humans is encoded by the SENP1 gene.

Sentrin-specific protease 6 is an enzyme that in humans is encoded by the SENP6 gene.

Ubiquitin carboxyl-terminal hydrolase or Ubiquitin specific protease 11 is an enzyme that in humans is encoded by the USP11 gene. USP11 belongs to the Ubiquitin specific proteases family (USPs) which is a sub-family of the Deubiquitinating enzymes (DUBs).USPs are multiple domain proteases and belong to the C19 cysteine proteases sub‒family. Depending on their domain architecture and position there is different homology between the various members. Generally the largest domain is the catalytic domain which harbours the three residue catalytic triad that is included inside conserved motifs. The catalytic domain also contains sequences that are not related with the catalysis function and their role is mostly not clearly understood at present, the length of these sequences varies for each USP and therefore the length of the whole catalytic domain can range from approximately 295 to 850 amino acids. Particular sequences inside the catalytic domain or at the N‒terminus of some USPs have been characterised as UBL and DUSP domains respectively. In some cases, regarding the UBL domains, it has been reported to have a catalysis enhancing function as in the case of USP7. In addition, a so‒called DU domain module is the combination of a DUSP domain followed by a UBL domain separated by a linker and is found in USP11 as well as in USP15 and USP4.

Sentrin-specific protease 2 is an enzyme that in humans is encoded by the SENP2 gene.

Ubiquitin carboxyl-terminal hydrolase 20 is an enzyme that in humans is encoded by the USP20 gene.

Sentrin-specific protease 8 is an enzyme that in humans is encoded by the SENP8 gene.
Ubiquitin-like 1-activating enzyme E1B (UBLE1B) also known as SUMO-activating enzyme subunit 2 (SAE2) is an enzyme that in humans is encoded by the UBA2 gene.
An isopeptidase is a protease enzyme that hydrolyzes isopeptide bonds, or amide bonds that occur outside the main chain in a polypeptide chain.

Ubiquitin-like proteins (UBLs) are a family of small proteins involved in post-translational modification of other proteins in a cell, usually with a regulatory function. The UBL protein family derives its name from the first member of the class to be discovered, ubiquitin (Ub), best known for its role in regulating protein degradation through covalent modification of other proteins. Following the discovery of ubiquitin, many additional evolutionarily related members of the group were described, involving parallel regulatory processes and similar chemistry. UBLs are involved in a widely varying array of cellular functions including autophagy, protein trafficking, inflammation and immune responses, transcription, DNA repair, RNA splicing, and cellular differentiation.
References
- ↑ Lima CD, Rawlings ND, Woessner JF (2004). "Ulp1 endopeptidase". In Barrett AJ (ed.). Handbook of Proteolytic Enzymes (2nd ed.). London: Elsevier. pp. 1340–1344.
- ↑ Li SJ, Hochstrasser M (March 1999). "A new protease required for cell-cycle progression in yeast". Nature. 398 (6724): 246–51. doi:10.1038/18457. PMID 10094048.
- ↑ Taylor DL, Ho JC, Oliver A, Watts FZ (March 2002). "Cell-cycle-dependent localisation of Ulp1, a Schizosaccharomyces pombe Pmt3 (SUMO)-specific protease". Journal of Cell Science. 115 (Pt 6): 1113–22. PMID 11884512.
- ↑ Li SJ, Hochstrasser M (March 2003). "The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity". The Journal of Cell Biology. 160 (7): 1069–81. doi:10.1083/jcb.200212052. PMC 2172760 . PMID 12654900.
- ↑ Ihara M, Koyama H, Uchimura Y, Saitoh H, Kikuchi A (June 2007). "Noncovalent binding of small ubiquitin-related modifier (SUMO) protease to SUMO is necessary for enzymatic activities and cell growth". The Journal of Biological Chemistry. 282 (22): 16465–75. doi: 10.1074/jbc.M610723200 . PMID 17428805.
- ↑ Mukhopadhyay D, Dasso M (June 2007). "Modification in reverse: the SUMO proteases". Trends in Biochemical Sciences. 32 (6): 286–95. doi:10.1016/j.tibs.2007.05.002. PMID 17499995.