Golden rice is a variety of rice produced through genetic engineering to biosynthesize beta-carotene, a precursor of vitamin A, in the edible parts of the rice. It is intended to produce a fortified food to be grown and consumed in areas with a shortage of dietary vitamin A. Vitamin A deficiency causes xerophthalmia, a range of eye conditions from night blindness to more severe clinical outcomes such as keratomalacia and corneal scars, and permanent blindness. Additionally, vitamin A deficiency also increases risk of mortality from measles and diarrhea in children. In 2013, the prevalence of deficiency was the highest in sub-Saharan Africa, and South Asia.
Carotenoids are yellow, orange, and red organic pigments that are produced by plants and algae, as well as several bacteria, archaea, and fungi. Carotenoids give the characteristic color to pumpkins, carrots, parsnips, corn, tomatoes, canaries, flamingos, salmon, lobster, shrimp, and daffodils. Over 1,100 identified carotenoids can be further categorized into two classes – xanthophylls and carotenes.

Carotenoid oxygenases are a family of enzymes involved in the cleavage of carotenoids to produce, for example, retinol, commonly known as vitamin A. This family includes an enzyme known as RPE65 which is abundantly expressed in the retinal pigment epithelium where it catalyzed the formation of 11-cis-retinol from all-trans-retinyl esters.
CRT is the gene cluster responsible for the biosynthesis of carotenoids. Those genes are found in eubacteria, in algae and are cryptic in Streptomyces griseus.
In enzymology, a carotene 7,8-desaturase (EC 1.14.99.30) is an enzyme that catalyzes the chemical reaction

Damascenones are a series of closely related chemical compounds that are components of a variety of essential oils. The damascenones belong to a family of chemicals known as rose ketones, which also includes damascones and ionones. beta-Damascenone is a major contributor to the aroma of roses, despite its very low concentration, and is an important fragrance chemical used in perfumery.

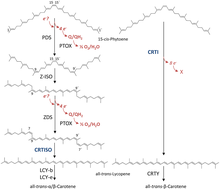
Phytoene is a 40-carbon intermediate in the biosynthesis of carotenoids. The synthesis of phytoene is the first committed step in the synthesis of carotenoids in plants. Phytoene is produced from two molecules of geranylgeranyl pyrophosphate (GGPP) by the action of the enzyme phytoene synthase. The two GGPP molecules are condensed together followed by removal of diphosphate and proton shift leading to the formation of phytoene.
Phytoene synthase is a transferase enzyme involved in the biosynthesis of carotenoids. It catalyzes the conversion of geranylgeranyl pyrophosphate to phytoene. This enzyme catalyses the following chemical reaction

15-cis-phytoene desaturases, are enzymes involved in the carotenoid biosynthesis in plants and cyanobacteria. Phytoene desaturases are membrane-bound enzymes localized in plastids and introduce two double bonds into their colorless substrate phytoene by dehydrogenation and isomerize two additional double bonds. This reaction starts a biochemical pathway involving three further enzymes called the poly-cis pathway and leads to the red colored lycopene. The homologous phytoene desaturase found in bacteria and fungi (CrtI) converts phytoene directly to lycopene by an all-trans pathway.
9,9'-dicis-zeta-carotene desaturase is an enzyme with systematic name 9,9'-dicis-zeta-corotene:quinone oxidoreductase. This enzyme catalyses the following chemical reaction
4,4'-Diapophytoene desaturase is an enzyme with systematic name 15-cis-4,4'-diapophytoene:FAD oxidoreductase. This enzyme catalyses the following chemical reaction
All-trans-zeta-carotene desaturase is an enzyme with systematic name all-trans-zeta-carotene:acceptor oxidoreductase. This enzyme catalyses the following chemical reaction
1-Hydroxycarotenoid 3,4-desaturase is an enzyme with systematic name 1-hydroxy-1,2-dihydrolycopene:acceptor oxidoreductase. This enzyme catalyses the following chemical reaction
Phytoene desaturase (neurosporene-forming) is an enzyme with systematic name 15-cis-phytoene:acceptor oxidoreductase (neurosporene-forming). This enzyme catalyses the following chemical reaction
Phytoene desaturase (zeta-carotene-forming) is an enzyme with systematic name 15-cis-phytoene:acceptor oxidoreductase (zeta-carotene-forming). This enzyme catalyses the following chemical reaction
Phytoene desaturase (3,4-didehydrolycopene-forming) is an enzyme with systematic name 15-cis-phytoene:acceptor oxidoreductase (3,4-didehydrolycopene-forming). This enzyme catalyses the following chemical reaction
Prolycopene isomerase is an enzyme with systematic name 7,9,7',9'-tetracis-lycopene cis-trans-isomerase. This enzyme catalyses the following chemical reaction
Beta-carotene isomerase is an enzyme with systematic name beta-carotene 9-cis-all-trans isomerase. This enzyme catalyses the following chemical reaction
Lycopene β-cyclase is an enzyme with systematic name carotenoid beta-end group lyase (decyclizing). This enzyme catalyses the following chemical reaction
Phytoene desaturase may refer to: