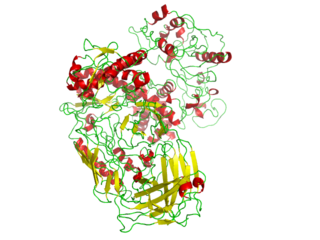
A polymerase is an enzyme that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication.
The SRC Homology 3 Domain is a small protein domain of about 60 amino acid residues. Initially, SH3 was described as a conserved sequence in the viral adaptor protein v-Crk. This domain is also present in the molecules of phospholipase and several cytoplasmic tyrosine kinases such as Abl and Src. It has also been identified in several other protein families such as: PI3 Kinase, Ras GTPase-activating protein, CDC24 and cdc25. SH3 domains are found in proteins of signaling pathways regulating the cytoskeleton, the Ras protein, and the Src kinase and many others. The SH3 proteins interact with adaptor proteins and tyrosine kinases. Interacting with tyrosine kinases, SH3 proteins usually bind far away from the active site. Approximately 300 SH3 domains are found in proteins encoded in the human genome. In addition to that, the SH3 domain was responsible for controlling protein-protein interactions in the signal transduction pathways and regulating the interactions of proteins involved in the cytoplasmic signaling.
The Enzyme Commission number is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze. As a system of enzyme nomenclature, every EC number is associated with a recommended name for the corresponding enzyme-catalyzed reaction.
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

The SH2domain is a structurally conserved protein domain contained within the Src oncoprotein and in many other intracellular signal-transducing proteins. SH2 domains allow proteins containing those domains to dock to phosphorylated tyrosine residues on other proteins. SH2 domains are commonly found in adaptor proteins that aid in the signal transduction of receptor tyrosine kinase pathways.

Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change the linking number of circular DNA by ±2. Topoisomerases are ubiquitous enzymes, found in all living organisms.
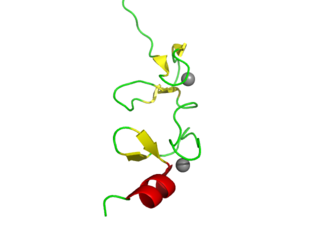
LIM domains are protein structural domains, composed of two contiguous zinc fingers, separated by a two-amino acid residue hydrophobic linker. The domain name is an acronym of the three genes in which it was first identified. LIM is a protein interaction domain that is involved in binding to many structurally and functionally diverse partners. The LIM domain appeared in eukaryotes sometime prior to the most recent common ancestor of plants, fungi, amoeba and animals. In animal cells, LIM domain-containing proteins often shuttle between the cell nucleus where they can regulate gene expression, and the cytoplasm where they are usually associated with actin cytoskeletal structures involved in connecting cells together and to the surrounding matrix, such as stress fibers, focal adhesions and adherens junctions.

A protein domain is a region of the protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains. One domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins.

Cytoplasmic protein NCK2 is a protein that in humans is encoded by the NCK2 gene.
Z-DNA binding protein 1, also known as Zuotin, is a Saccharomyces cerevisiae yeast gene.

A Per-Arnt-Sim (PAS) domain is a protein domain found in all kingdoms of life. Generally, the PAS domain acts as a molecular sensor, whereby small molecules and other proteins associate via binding of the PAS domain. Due to this sensing capability, the PAS domain has been shown as the key structural motif involved in protein-protein interactions of the circadian clock, and it is also a common motif found in signaling proteins, where it functions as a signaling sensor.
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.
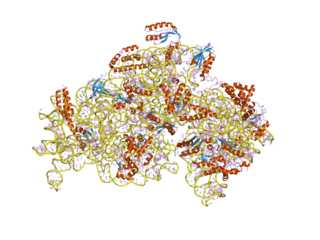
In molecular biology, S4 domain refers to a small RNA-binding protein domain found in a ribosomal protein named uS4. The S4 domain is approximately 60-65 amino acid residues long, occurs in a single copy at various positions in different proteins and was originally found in pseudouridine syntheses, a bacterial ribosome-associated protein.
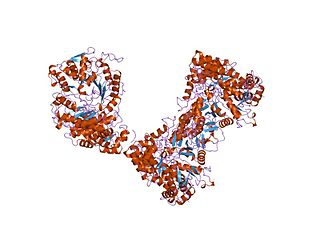
In molecular biology, the ATP-cone is an evolutionarily mobile, ATP-binding regulatory domain which is found in a variety of proteins including ribonucleotide reductases, phosphoglycerate kinases and transcriptional regulators.
In molecular biology, the cache domain is an extracellular protein domain that is predicted to have a role in small-molecule recognition in a wide range of proteins, including the animal dihydropyridine-sensitive voltage-gated Ca2+ channel alpha-2delta subunit, and various bacterial chemotaxis receptors. The name Cache comes from CAlcium channels and CHEmotaxis receptors. This domain consists of an N-terminal part with three predicted strands and an alpha-helix, and a C-terminal part with a strand dyad followed by a relatively unstructured region. The N-terminal portion of the (unpermuted) Cache domain contains three predicted strands that could form a sheet analogous to that present in the core of the PAS domain structure. Cache domains are particularly widespread in bacteria such as Vibrio cholerae. The animal calcium channel alpha-2delta subunits might have acquired a part of their extracellular domains from a bacterial source. The Cache domain appears to have arisen from the GAF-PAS fold despite their divergent functions.
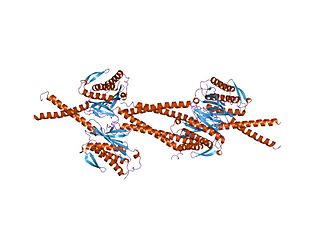
In molecular biology, the HORMA domain is a protein domain that has been suggested to recognise chromatin states resulting from DNA adducts, double stranded breaks or non-attachment to the spindle and act as an adaptor that recruits other proteins. Hop1 is a meiosis-specific protein, Rev7 is required for DNA damage induced mutagenesis, and MAD2 is a spindle checkpoint protein which prevents progression of the cell cycle upon detection of a defect in mitotic spindle integrity.

In molecular biology, the KilA-N domain is a conserved protein domain.
A protein superfamily is the largest grouping (clade) of proteins for which common ancestry can be inferred. Usually this common ancestry is inferred from structural alignment and mechanistic similarity, even if no sequence similarity is evident. Sequence homology can then be deduced even if not apparent. Superfamilies typically contain several protein families which show sequence similarity within each family. The term protein clan is commonly used for protease and glycosyl hydrolases superfamilies based on the MEROPS and CAZy classification systems.

PrimPol is a protein encoded by the PRIMPOL gene in humans. PrimPol is a eukaryotic protein with both DNA polymerase and DNA Primase activities involved in translesion DNA synthesis. It is the first eukaryotic protein to be identified with priming activity using deoxyribonucleotides. It is also the first protein identified in the mitochondria to have translesion DNA synthesis activities.

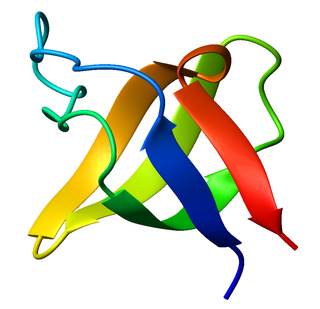
The jelly roll or Swiss roll fold is a protein fold or supersecondary structure composed of eight beta strands arranged in two four-stranded sheets. The name of the structure was introduced by Jane S. Richardson in 1981, reflecting its resemblance to the jelly or Swiss roll cake. The fold is an elaboration on the Greek key motif and is sometimes considered a form of beta barrel. It is very common in viral proteins, particularly viral capsid proteins. Taken together, the jelly roll and Greek key structures comprise around 30% of the all-beta proteins annotated in the Structural Classification of Proteins (SCOP) database.














