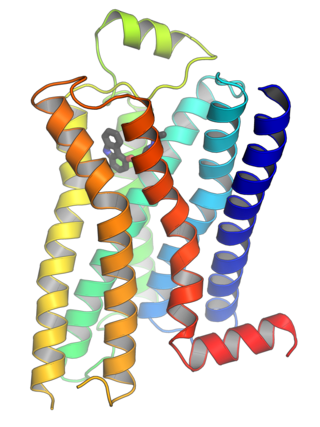
G protein-coupled receptors (GPCRs), also known as seven-(pass)-transmembrane domain receptors, 7TM receptors, heptahelical receptors, serpentine receptors, and G protein-linked receptors (GPLR), form a large group of evolutionarily related proteins that are cell surface receptors that detect molecules outside the cell and activate cellular responses. They are coupled with G proteins. They pass through the cell membrane seven times in the form of six loops of amino acid residues, which is why they are sometimes referred to as seven-transmembrane receptors. Ligands can bind either to the extracellular N-terminus and loops or to the binding site within transmembrane helices. They are all activated by agonists, although a spontaneous auto-activation of an empty receptor has also been observed.

Peripheral membrane proteins, or extrinsic membrane proteins, are membrane proteins that adhere only temporarily to the biological membrane with which they are associated. These proteins attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins.
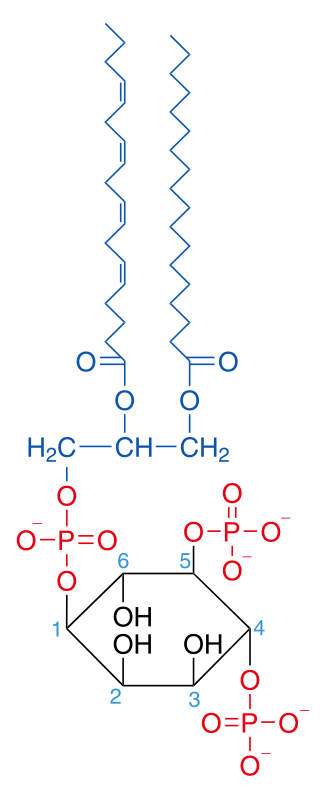
Phosphatidylinositol 4,5-bisphosphate or PtdIns(4,5)P2, also known simply as PIP2 or PI(4,5)P2, is a minor phospholipid component of cell membranes. PtdIns(4,5)P2 is enriched at the plasma membrane where it is a substrate for a number of important signaling proteins. PIP2 also forms lipid clusters that sort proteins.

Pleckstrin homology domain or (PHIP) is a protein domain of approximately 120 amino acids that occurs in a wide range of proteins involved in intracellular signaling or as constituents of the cytoskeleton.
The epsin N-terminal homology (ENTH) domain is a structural domain that is found in proteins involved in endocytosis and cytoskeletal machinery.

A C2 domain is a protein structural domain involved in targeting proteins to cell membranes. The typical version (PKC-C2) has a beta-sandwich composed of 8 β-strands that co-ordinates two or three calcium ions, which bind in a cavity formed by the first and final loops of the domain, on the membrane binding face. Many other C2 domain families don't have calcium binding activity.
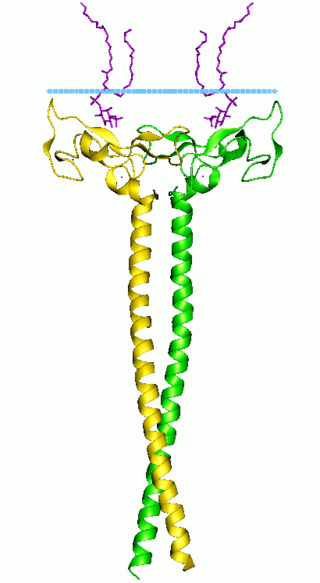
In molecular biology the FYVE zinc finger domain is named after the four cysteine-rich proteins: Fab 1, YOTB, Vac 1, and EEA1, in which it has been found. FYVE domains bind phosphatidylinositol 3-phosphate, in a way dependent on its metal ion coordination and basic amino acids. The FYVE domain inserts into cell membranes in a pH-dependent manner. The FYVE domain has been connected to vacuolar protein sorting and endosome function.
Cell wall protein 2 (CWP2) is a cell wall protein, produced by Saccharomyces cerevisiae and Saccharomyces pastorianus. It occurs throughout the cell wall and has close homology with the CWP1 gene.
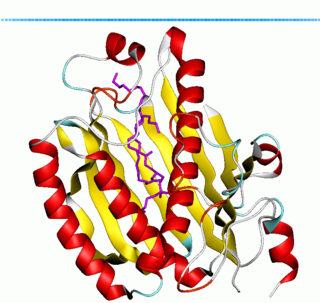
Phosphatidylinositol transfer protein (PITP) or priming in exocytosis protein 3 (PEP3) is a ubiquitous cytosolic domain involved in transport of phospholipids from their site of synthesis in the endoplasmic reticulum and Golgi to other cell membranes.
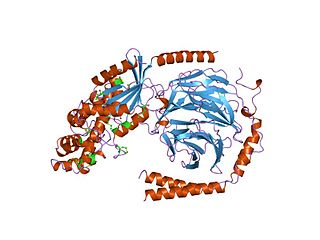
G alpha subunits are one of the three types of subunit of guanine nucleotide binding proteins, which are membrane-associated, heterotrimeric G proteins.

Regulators of G protein signaling (RGS) are protein structural domains or the proteins that contain these domains, that function to activate the GTPase activity of heterotrimeric G-protein α-subunits.

SEC14L2 is a gene that, in humans, encodes the protein SEC14-like protein 2.

Phosphatidylinositol 4-kinase beta is an enzyme that in humans is encoded by the PI4KB gene.
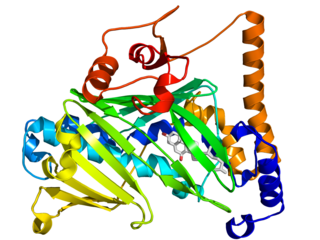
The oxysterol-binding protein (OSBP)-related proteins (ORPs) are a family of lipid transfer proteins (LTPs). Concretely, they constitute a family of sterol and phosphoinositide binding and transfer proteins in eukaryotes that are conserved from yeast to humans. They are lipid-binding proteins implicated in many cellular processes related with oxysterol, including signaling, vesicular trafficking, lipid metabolism, and nonvesicular sterol transport.
Membrane contact sites (MCS) are close appositions between two organelles. Ultrastructural studies typically reveal an intermembrane distance in the order of the size of a single protein, as small as 10 nm or wider, with no clear upper limit. These zones of apposition are highly conserved in evolution. These sites are thought to be important to facilitate signalling, and they promote the passage of small molecules, including ions, lipids and reactive oxygen species. MCS are important in the function of the endoplasmic reticulum (ER), since this is the major site of lipid synthesis within cells. The ER makes close contact with many organelles, including mitochondria, Golgi, endosomes, lysosomes, peroxisomes, chloroplasts and the plasma membrane. Both mitochondria and sorting endosomes undergo major rearrangements leading to fission where they contact the ER. Sites of close apposition can also form between most of these organelles most pairwise combinations. First mentions of these contact sites can be found in papers published in the late 1950s mainly visualized using electron microscopy (EM) techniques. Copeland and Dalton described them as “highly specialized tubular form of endoplasmic reticulum in association with the mitochondria and apparently in turn, with the vascular border of the cell”.
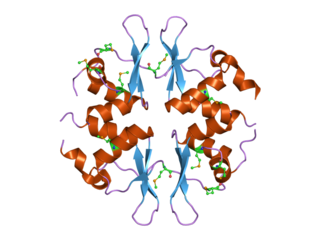
In molecular biology, the CBS domain is a protein domain found in a range of proteins in all species from bacteria to humans. It was first identified as a conserved sequence region in 1997 and named after cystathionine beta synthase, one of the proteins it is found in. CBS domains are also found in a wide variety of other proteins such as inosine monophosphate dehydrogenase, voltage gated chloride channels and AMP-activated protein kinase (AMPK). CBS domains regulate the activity of associated enzymatic and transporter domains in response to binding molecules with adenosyl groups such as AMP and ATP, or s-adenosylmethionine.

Sec14 is a cytosolic protein found in yeast which plays a role in the regulation of several cellular functions, specifically those related to intracellular transport. Encoded by the Sec14 gene, Sec14p may transport phosphatidylinositol and phosphatidylcholine produced in the endoplasmic reticulum and the Golgi body to other cellular membranes. Additionally, Sec14p potentially plays a role in the localization of lipid raft proteins. Sec14p is an essential gene in yeast, and is homologous in function to phosphatidylinositol transfer protein in mammals. A conditional mutant with non-functional Sec14p presents with Berkeley bodies and deficiencies in protein secretion.

In molecular biology, the cyclase-associated protein family (CAP) is a family of highly conserved actin-binding proteins present in a wide range of organisms including yeast, flies, plants, and mammals. CAPs are multifunctional proteins that contain several structural domains. CAP is involved in species-specific signalling pathways. In Drosophila, CAP functions in Hedgehog-mediated eye development and in establishing oocyte polarity. In Dictyostelium discoideum, CAP is involved in microfilament reorganisation near the plasma membrane in a PIP2-regulated manner and is required to perpetuate the cAMP relay signal to organise fruitbody formation. In plants, CAP is involved in plant signalling pathways required for co-ordinated organ expansion. In yeast, CAP is involved in adenylate cyclase activation, as well as in vesicle trafficking and endocytosis. In both yeast and mammals, CAPs appear to be involved in recycling G-actin monomers from ADF/cofilins for subsequent rounds of filament assembly. In mammals, there are two different CAPs that share 64% amino acid identity.
A FFAT motif is a protein sequence motif of six defined amino acids plus neighbouring residues that binds to proteins in the VAP protein family.
Nir1 or membrane-associated phosphatidylinositol transfer protein 3 (PITPNM3) is a mammalian protein that localizes to endoplasmic reticulum (ER) and plasma membrane (PM) membrane contact sites (MCS) and aids the transfer of phosphatidylinositol between these two membranes, potentially by recruiting additional proteins to the ER-PM MCS. It is encoded by the gene PITPNM3.