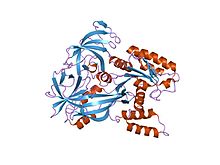
GTPases are a large family of hydrolase enzymes that bind to the nucleotide guanosine triphosphate (GTP) and hydrolyze it to guanosine diphosphate (GDP). The GTP binding and hydrolysis takes place in the highly conserved P-loop "G domain", a protein domain common to many GTPases.

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The Total result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
Initiation factors are proteins that bind to the small subunit of the ribosome during the initiation of translation, a part of protein biosynthesis.

Elongation factors are a set of proteins that function at the ribosome, during protein synthesis, to facilitate translational elongation from the formation of the first to the last peptide bond of a growing polypeptide. Most common elongation factors in prokaryotes are EF-Tu, EF-Ts, EF-G. Bacteria and eukaryotes use elongation factors that are largely homologous to each other, but with distinct structures and different research nomenclatures.

A DNA clamp, also known as a sliding clamp or β-clamp, is a protein complex that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because one of the rate-limiting steps in the DNA synthesis reaction is the association of the polymerase with the DNA template, the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.

EF-Tu is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochrondria as TUFM.
eEF-1 are two eukaryotic elongation factors. It forms two complexes, the EF-Tu homolog EF-1A and the EF-Ts homolog EF-1B, the former's guanide exchange factor. Both are also found in archaea.

Elongation factor 1-delta is a protein that in humans is encoded by the EEF1D gene.

Elongation factor 1-alpha 1 (eEF1a1) is a translation elongation protein, expressed across eukaryotes. In humans, it is encoded by the EEF1A1 gene.

Elongation factor 1-beta is a protein that in humans is encoded by the EEF1B2 gene.
eIF2B is a protein complex found in eukaryotes. It is the guanine nucleotide exchange factor for the eukaryotic initiation factor 2 and therefore converts the inactive eIF2-GDP to the active eIF2-GTP. This activation is hindered by phosphorylation of the alpha subunit of eIF2, which leads to a stable eIF2α-P-GDP-eIF2B complex and therefore inhibits translation initiation.
Eukaryotic Initiation Factor 2 (eIF2) is a eukaryotic initiation factor. It is required for most forms of eukaryotic translation initiation. eIF2 mediates the binding of tRNAiMet to the ribosome in a GTP-dependent manner. eIF2 is a heterotrimer consisting of an alpha, a beta, and a gamma subunit.
EF-Ts is one of the prokaryotic elongation factors. It is found in human mitochrondria as TSFM. It is similar to eukaryotic EF-1B.

EF-P is an essential protein that in bacteria stimulates the formation of the first peptide bonds in protein synthesis. Studies show that EF-P prevents ribosomes from stalling during the synthesis of proteins containing consecutive prolines. EF-P binds to a site located between the binding site for the peptidyl tRNA and the exiting tRNA. It spans both ribosomal subunits with its amino-terminal domain positioned adjacent to the aminoacyl acceptor stem and its carboxyl-terminal domain positioned next to the anticodon stem-loop of the P site-bound initiator tRNA. The EF-P protein shape and size is very similar to a tRNA and interacts with the ribosome via the exit “E” site on the 30S subunit and the peptidyl-transferase center (PTC) of the 50S subunit. EF-P is a translation aspect of an unknown function, therefore It probably functions indirectly by altering the affinity of the ribosome for aminoacyl-tRNA, thus increasing their reactivity as acceptors for peptidyl transferase.
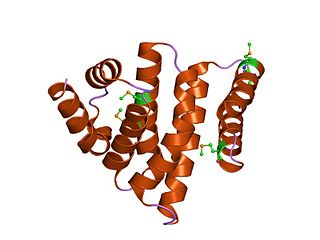
In molecular biology, the protein domain eIF4-gamma/eIF5/eIF2-epsilon is a family of evolutionarily related proteins. This domain is found at the C-terminus of several translation Initiation factors. It was first detected at the very C-termini of the yeast protein GCD6, eIF-2B epsilon, and two other eukaryotic translation initiation factors, eIF-4 gamma and eIF-5 and it may be involved in the interaction of eIF-2B, eIF-4 gamma, and eIF-5 with eIF-2.
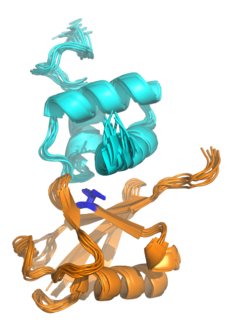
Ubiquitin-associated (UBA) domains are protein domains that non-covalently interact with ubiquitin through protein-protein interactions. Ubiquitin is a small protein that is covalently linked to other proteins as part of intracellular signaling pathways, often as a signal for protein degradation. UBA domains are among the most common ubiquitin-binding domains.
Archaeal translation is the process by which messenger RNA is translated into proteins in archaea. Not much is known on this subject, but on the protein level it seems to resemble eukaryotic translation.