Related Research Articles

The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA and some of its organelles, and subsequently the partitioning of its cytoplasm, chromosomes and other components into two daughter cells in a process called cell division.

Mitosis is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis is an equational division which gives rise to genetically identical cells in which the total number of chromosomes is maintained. Mitosis is preceded by the S phase of interphase and is followed by telophase and cytokinesis, which divide the cytoplasm, organelles, and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis altogether define the mitotic phase of a cell cycle—the division of the mother cell into two daughter cells genetically identical to each other.

Cell division is the process by which a parent cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there are two distinct types of cell division: a vegetative division (mitosis), producing daughter cells genetically identical to the parent cell, and a cell division that produces haploid gametes for sexual reproduction (meiosis), reducing the number of chromosomes from two of each type in the diploid parent cell to one of each type in the daughter cells. Mitosis is a part of the cell cycle, in which, replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. In general, mitosis is preceded by the S stage of interphase and is followed by telophase and cytokinesis; which divides the cytoplasm, organelles, and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of mitosis all together define the M phase of an animal cell cycle—the division of the mother cell into two genetically identical daughter cells. To ensure proper progression through the cell cycle, DNA damage is detected and repaired at various checkpoints throughout the cycle. These checkpoints can halt progression through the cell cycle by inhibiting certain cyclin-CDK complexes. Meiosis undergoes two divisions resulting in four haploid daughter cells. Homologous chromosomes are separated in the first division of meiosis, such that each daughter cell has one copy of each chromosome. These chromosomes have already been replicated and have two sister chromatids which are then separated during the second division of meiosis. Both of these cell division cycles are used in the process of sexual reproduction at some point in their life cycle. Both are believed to be present in the last eukaryotic common ancestor.

The G1 phase, gap 1 phase, or growth 1 phase, is the first of four phases of the cell cycle that takes place in eukaryotic cell division. In this part of interphase, the cell synthesizes mRNA and proteins in preparation for subsequent steps leading to mitosis. G1 phase ends when the cell moves into the S phase of interphase. Around 30 to 40 percent of cell cycle time is spent in the G1 phase.

Telophase is the final stage in both meiosis and mitosis in a eukaryotic cell. During telophase, the effects of prophase and prometaphase are reversed. As chromosomes reach the cell poles, a nuclear envelope is re-assembled around each set of chromatids, the nucleoli reappear, and chromosomes begin to decondense back into the expanded chromatin that is present during interphase. The mitotic spindle is disassembled and remaining spindle microtubules are depolymerized. Telophase accounts for approximately 2% of the cell cycle's duration.
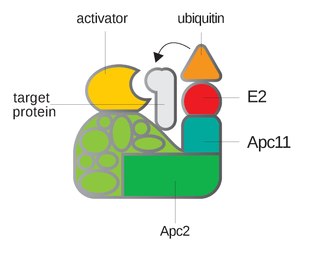
Anaphase-promoting complex is an E3 ubiquitin ligase that marks target cell cycle proteins for degradation by the 26S proteasome. The APC/C is a large complex of 11–13 subunit proteins, including a cullin (Apc2) and RING (Apc11) subunit much like SCF. Other parts of the APC/C have unknown functions but are highly conserved.

Nocodazole is an antineoplastic agent which exerts its effect in cells by interfering with the polymerization of microtubules. Microtubules are one type of fibre which constitutes the cytoskeleton, and the dynamic microtubule network has several important roles in the cell, including vesicular transport, forming the mitotic spindle and in cytokinesis. Several drugs including vincristine and colcemid are similar to nocodazole in that they interfere with microtubule polymerization.
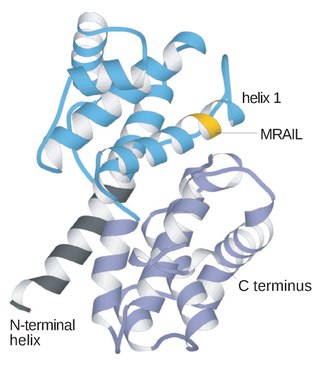
Cyclins are proteins that control the progression of a cell through the cell cycle by activating cyclin-dependent kinases (CDK).
Maturation-promoting factor (abbreviated MPF, also called mitosis-promoting factor or M-Phase-promoting factor) is the cyclin–Cdk complex that was discovered first in frog eggs. It stimulates the mitotic and meiotic phases of the cell cycle. MPF promotes the entrance into mitosis (the M phase) from the G2 phase by phosphorylating multiple proteins needed during mitosis. MPF is activated at the end of G2 by a phosphatase, which removes an inhibitory phosphate group added earlier.

The spindle checkpoint, also known as the metaphase-to-anaphase transition, the spindle assembly checkpoint (SAC), the metaphase checkpoint, or the mitotic checkpoint, is a cell cycle checkpoint during metaphase of mitosis or meiosis that prevents the separation of the duplicated chromosomes (anaphase) until each chromosome is properly attached to the spindle. To achieve proper segregation, the two kinetochores on the sister chromatids must be attached to opposite spindle poles. Only this pattern of attachment will ensure that each daughter cell receives one copy of the chromosome. The defining biochemical feature of this checkpoint is the stimulation of the anaphase-promoting complex by M-phase cyclin-CDK complexes, which in turn causes the proteolytic destruction of cyclins and proteins that hold the sister chromatids together.

G2 phase, Gap 2 phase, or Growth 2 phase, is the third subphase of interphase in the cell cycle directly preceding mitosis. It follows the successful completion of S phase, during which the cell’s DNA is replicated. G2 phase ends with the onset of prophase, the first phase of mitosis in which the cell’s chromatin condenses into chromosomes.
A spindle poison, also known as a spindle toxin, is a poison that disrupts cell division by affecting the protein threads that connect the centromere regions of chromosomes, known as spindles. Spindle poisons effectively cease the production of new cells by interrupting the mitosis phase of cell division at the spindle assembly checkpoint (SAC). However, as numerous and varied as they are, spindle poisons are not yet 100% effective at ending the formation of tumors (neoplasms). Although not 100% effective, substantive therapeutic efficacy has been found in these types of chemotherapeutic treatments. The mitotic spindle is composed of microtubules that aid, along with regulatory proteins, each other in the activity of appropriately segregating replicated chromosomes. Certain compounds affecting the mitotic spindle have proven highly effective against solid tumors and hematological malignancies.

Cell cycle checkpoints are control mechanisms in the eukaryotic cell cycle which ensure its proper progression. Each checkpoint serves as a potential termination point along the cell cycle, during which the conditions of the cell are assessed, with progression through the various phases of the cell cycle occurring only when favorable conditions are met. There are many checkpoints in the cell cycle, but the three major ones are: the G1 checkpoint, also known as the Start or restriction checkpoint or Major Checkpoint; the G2/M checkpoint; and the metaphase-to-anaphase transition, also known as the spindle checkpoint. Progression through these checkpoints is largely determined by the activation of cyclin-dependent kinases by regulatory protein subunits called cyclins, different forms of which are produced at each stage of the cell cycle to control the specific events that occur therein.
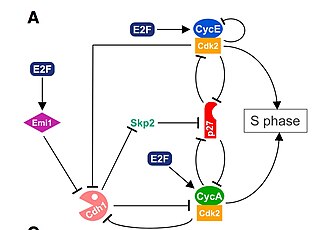
The G1/S transition is a stage in the cell cycle at the boundary between the G1 phase, in which the cell grows, and the S phase, during which DNA is replicated. It is governed by cell cycle checkpoints to ensure cell cycle integrity and the subsequent S phase can pause in response to improperly or partially replicated DNA. During this transition the cell makes decisions to become quiescent, differentiate, make DNA repairs, or proliferate based on environmental cues and molecular signaling inputs. The G1/S transition occurs late in G1 and the absence or improper application of this highly regulated checkpoint can lead to cellular transformation and disease states such as cancer.
Cyclin A is a member of the cyclin family, a group of proteins that function in regulating progression through the cell cycle. The stages that a cell passes through that culminate in its division and replication are collectively known as the cell cycle Since the successful division and replication of a cell is essential for its survival, the cell cycle is tightly regulated by several components to ensure the efficient and error-free progression through the cell cycle. One such regulatory component is cyclin A which plays a role in the regulation of two different cell cycle stages.

Geminin, DNA replication inhibitor, also known as GMNN, is a protein in humans encoded by the GMNN gene. A nuclear protein present in most eukaryotes and highly conserved across species, numerous functions have been elucidated for geminin including roles in metazoan cell cycle, cellular proliferation, cell lineage commitment, and neural differentiation. One example of its function is the inhibition of Cdt1.
A series of biochemical switches control transitions between and within the various phases of the cell cycle. The cell cycle is a series of complex, ordered, sequential events that control how a single cell divides into two cells, and involves several different phases. The phases include the G1 and G2 phases, DNA replication or S phase, and the actual process of cell division, mitosis or M phase. During the M phase, the chromosomes separate and cytokinesis occurs.
Cell cycle analysis by DNA content measurement is a method that most frequently employs flow cytometry to distinguish cells in different phases of the cell cycle. Before analysis, the cells are usually permeabilised and treated with a fluorescent dye that stains DNA quantitatively, such as propidium iodide (PI) or 4,6-diamidino-2-phenylindole (DAPI). The fluorescence intensity of the stained cells correlates with the amount of DNA they contain. As the DNA content doubles during the S phase, the DNA content (and thereby intensity of fluorescence) of cells in the G0 phase and G1 phase (before S), in the S phase, and in the G2 phase and M phase (after S) identifies the cell cycle phase position in the major phases (G0/G1 versus S versus G2/M phase) of the cell cycle. The cellular DNA content of individual cells is often plotted as their frequency histogram to provide information about relative frequency (percentage) of cells in the major phases of the cell cycle.
Mitotic exit is an important transition point that signifies the end of mitosis and the onset of new G1 phase for a cell, and the cell needs to rely on specific control mechanisms to ensure that once it exits mitosis, it never returns to mitosis until it has gone through G1, S, and G2 phases and passed all the necessary checkpoints. Many factors including cyclins, cyclin-dependent kinases (CDKs), ubiquitin ligases, inhibitors of cyclin-dependent kinases, and reversible phosphorylations regulate mitotic exit to ensure that cell cycle events occur in correct order with fewest errors. The end of mitosis is characterized by spindle breakdown, shortened kinetochore microtubules, and pronounced outgrowth of astral (non-kinetochore) microtubules. For a normal eukaryotic cell, mitotic exit is irreversible.
Induced cell cycle arrest is the use of a chemical or genetic manipulation to artificially halt progression through the cell cycle. Cellular processes like genome duplication and cell division stop. It can be temporary or permanent. It is an artificial activation of naturally occurring cell cycle checkpoints, induced by exogenous stimuli controlled by an experimenter.
References
- ↑ Keng, PC (September 1980). "Synchronization of 9L rat brain tumor cells by centrifugal elutriation". Cell Biophysics. 2 (3): 191–206. doi:10.1007/BF02790449. PMID 6159093. S2CID 7679022.
- ↑ Krek, Wilhelm (1995). Methods in Enzymology. Elsevier Inc. pp. 114–124.
- 1 2 3 Banfalvi G. (2011) Overview of Cell Synchronization. In: Banfalvi G. (eds) Cell Cycle Synchronization. Methods in Molecular Biology (Methods and Protocols), vol 761. Humana Press.
- ↑ Coquelle, A; et al. (30 November 2006). "Enrichment of non-synchronized cells in the G1, S and G2 phases of the cell cycle for the study of apoptosis". Biochemical Pharmacology. 72 (11): 1396–1404. doi:10.1016/j.bcp.2006.04.014. PMID 16765323.
- ↑ Zieve, Gary W.; Turnbull, Deborah; Mullins, J.Michael; McIntosh, J.Richard (April 1980). "Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor Nocodazole: Nocodazole accumulated mitotic cells". Experimental Cell Research. 126 (2): 397–405. doi:10.1016/0014-4827(80)90279-7. PMID 6153987.
- ↑ Vassilev, Lyubomir T (8 November 2006). "Cell Cycle Synchronization at the G2/M Phase Border by Reversible Inhibition of CDK1". Cell Cycle. 5 (22): 2555–2556. doi: 10.4161/cc.5.22.3463 . PMID 17172841 – via Taylor & Francis Online.
- ↑ Azevedo, W. F.; Leclerc, S.; Meijer, L.; Havlicek, I.; Strnad, M.; Kim, S. H. (1997). "Inhibition of cyclin-dependent kinases by a purine analogs: crystal structure of human cdk2 complexed with roscovitine". Eur. J. Biochem. 243 (1–2): 518–526. doi: 10.1111/j.1432-1033.1997.0518a.x . PMID 9030780.
- ↑ G. Banfalvi (ed.), Cell Cycle Synchronization, Methods in Molecular Biology 761, DOI 10.1007/978-1-61779-182-6_10, © Springer Science+Business Media, LLC 2011
- ↑ Koc, Ahmet; Wheeler, Linda J.; Mathews, Christopher K.; Merrill, Gary F. (21 October 2003). "Hydroxyurea Arrests DNA Replication by a Mechanism That Preserves Basal dNTP Pools". Journal of Biological Chemistry. 279 (1): 223–230. doi: 10.1074/jbc.m303952200 . PMID 14573610.
- ↑ Nagano, H; Ikegami, S (November 1980). "Aphidicolin: a specific inhibitor of eukaryotic DNA polymerase alpha". Seikagaku: The Journal of Japanese Biomedical Society. 52 (11): 1208–1216. PMID 6790638.
- ↑ Sala, F.; Galli, M. G.; Levi, M.; Burroni, D.; Parisi, B.; Pedrali-Noy, G.; Spadari, S. (1981). "Functional roles of the plant alpha-like and gamma-like DNA polymerases". FEBS Lett. 124 (1): 112–118. doi:10.1016/0014-5793(81)80064-6. PMID 6783441. S2CID 85153639.
- ↑ Keyomarsi, Khandan; Sandoval, Larue; Band, Vilma; Pardee, Arthur B. (1 July 1991). "Synchronization of Tumor and Normal Cells from G1 to Multiple Cell Cycles by Lovastatin". Cancer Research. 51 (13): 3602–3609. PMID 1711413.
- ↑ "Mitotic cell selection". Biology Online Dictionary. 3 October 2005. Archived from the original on 2 April 2019. Retrieved 15 December 2018.
- 1 2 Davis, Penny K; Ho, Alan; Dowdy, Steven F (June 2001). "Biological Methods for Cell-Cycle Synchronization of Mammalian Cells". BioTechniques. 30 (3): 1322–1331. doi: 10.2144/01306rv01 . PMID 11414226.
- ↑ Zeng, Qi; Hong, Wanjin (11 March 2008). "The Emerging Role of the Hippo Pathway in Cell Contact Inhibition, Organ Size Control, and Cancer Development in Mammals". Cancer Cell. 13 (3): 188–192. doi: 10.1016/j.ccr.2008.02.011 . PMID 18328423.