Related Research Articles
Microevolution is the change in allele frequencies that occurs over time within a population. This change is due to four different processes: mutation, selection, gene flow and genetic drift. This change happens over a relatively short amount of time compared to the changes termed macroevolution.
Speciation is the evolutionary process by which populations evolve to become distinct species. The biologist Orator F. Cook coined the term in 1906 for cladogenesis, the splitting of lineages, as opposed to anagenesis, phyletic evolution within lineages. Charles Darwin was the first to describe the role of natural selection in speciation in his 1859 book On the Origin of Species. He also identified sexual selection as a likely mechanism, but found it problematic.

Domestication is a multi-generational mutualistic relationship in which an animal species, such as humans or leafcutter ants, takes over control and care of another species, such as sheep or fungi, to obtain from them a steady supply of resources, such as meat, milk, or labor. The process is gradual and geographically diffuse, based on trial and error.

A population bottleneck or genetic bottleneck is a sharp reduction in the size of a population due to environmental events such as famines, earthquakes, floods, fires, disease, and droughts; or human activities such as genocide, speciocide, widespread violence or intentional culling. Such events can reduce the variation in the gene pool of a population; thereafter, a smaller population, with a smaller genetic diversity, remains to pass on genes to future generations of offspring. Genetic diversity remains lower, increasing only when gene flow from another population occurs or very slowly increasing with time as random mutations occur. This results in a reduction in the robustness of the population and in its ability to adapt to and survive selecting environmental changes, such as climate change or a shift in available resources. Alternatively, if survivors of the bottleneck are the individuals with the greatest genetic fitness, the frequency of the fitter genes within the gene pool is increased, while the pool itself is reduced.

In population genetics, gene flow is the transfer of genetic material from one population to another. If the rate of gene flow is high enough, then two populations will have equivalent allele frequencies and therefore can be considered a single effective population. It has been shown that it takes only "one migrant per generation" to prevent populations from diverging due to drift. Populations can diverge due to selection even when they are exchanging alleles, if the selection pressure is strong enough. Gene flow is an important mechanism for transferring genetic diversity among populations. Migrants change the distribution of genetic diversity among populations, by modifying allele frequencies. High rates of gene flow can reduce the genetic differentiation between the two groups, increasing homogeneity. For this reason, gene flow has been thought to constrain speciation and prevent range expansion by combining the gene pools of the groups, thus preventing the development of differences in genetic variation that would have led to differentiation and adaptation. In some cases dispersal resulting in gene flow may also result in the addition of novel genetic variants under positive selection to the gene pool of a species or population
Allopatric speciation – also referred to as geographic speciation, vicariant speciation, or its earlier name the dumbbell model – is a mode of speciation that occurs when biological populations become geographically isolated from each other to an extent that prevents or interferes with gene flow.

In evolutionary biology, sympatric speciation is the evolution of a new species from a surviving ancestral species while both continue to inhabit the same geographic region. In evolutionary biology and biogeography, sympatric and sympatry are terms referring to organisms whose ranges overlap so that they occur together at least in some places. If these organisms are closely related, such a distribution may be the result of sympatric speciation. Etymologically, sympatry is derived from Greek συν (sun-) 'together', and πατρίς (patrís) 'fatherland'. The term was coined by Edward Bagnall Poulton in 1904, who explains the derivation.

The domestication of vertebrates is the mutual relationship between vertebrate animals including birds and mammals, and the humans who have influence on their care and reproduction.

Introgression, also known as introgressive hybridization, in genetics is the transfer of genetic material from one species into the gene pool of another by the repeated backcrossing of an interspecific hybrid with one of its parent species. Introgression is a long-term process, even when artificial; it may take many hybrid generations before significant backcrossing occurs. This process is distinct from most forms of gene flow in that it occurs between two populations of different species, rather than two populations of the same species.

The Ficedula flycatchers are a genus of Old World flycatchers. The genus is the largest in the family, containing around thirty species. They have sometimes been included in the genus Muscicapa. The genus is found in Europe, Asia and Africa. Several species are highly migratory, whereas other species are sedentary.
The mechanisms of reproductive isolation are a collection of evolutionary mechanisms, behaviors and physiological processes critical for speciation. They prevent members of different species from producing offspring, or ensure that any offspring are sterile. These barriers maintain the integrity of a species by reducing gene flow between related species.
In genetics, a selective sweep is the process through which a new beneficial mutation that increases its frequency and becomes fixed in the population leads to the reduction or elimination of genetic variation among nucleotide sequences that are near the mutation. In selective sweep, positive selection causes the new mutation to reach fixation so quickly that linked alleles can "hitchhike" and also become fixed.

Hybrid speciation is a form of speciation where hybridization between two different species leads to a new species, reproductively isolated from the parent species. Previously, reproductive isolation between two species and their parents was thought to be particularly difficult to achieve, and thus hybrid species were thought to be very rare. With DNA analysis becoming more accessible in the 1990s, hybrid speciation has been shown to be a somewhat common phenomenon, particularly in plants. In botanical nomenclature, a hybrid species is also called a nothospecies. Hybrid species are by their nature polyphyletic.
Population genomics is the large-scale comparison of DNA sequences of populations. Population genomics is a neologism that is associated with population genetics. Population genomics studies genome-wide effects to improve our understanding of microevolution so that we may learn the phylogenetic history and demography of a population.
A genetic isolate is a population of organisms that has little to no genetic mixing with other organisms of the same species due to geographic isolation or other factors that prevent reproduction. Genetic isolates form new species through an evolutionary process known as speciation. All modern species diversity is a product of genetic isolates and evolution.
Hybrizyme is a term coined to indicate novel or normally rare gene variants that are associated with hybrid zones, geographic areas where two related taxa meet, mate, and produce hybrid offspring. The hybrizyme phenomenon is widespread and these alleles occur commonly, if not in all hybrid zones. Initially considered to be caused by elevated rates of mutation in hybrids, the most probable hypothesis infers that they are the result of negative (purifying) selection. Namely, in the center of the hybrid zone, negative selection purges alleles against hybrid disadvantage. Stated differently, any allele that will decrease reproductive isolation is favored and any linked alleles also increase their frequency by genetic hitchhiking. If the linked alleles used to be rare variants in the parental taxa, they will become more common in the area where the hybrids are formed.
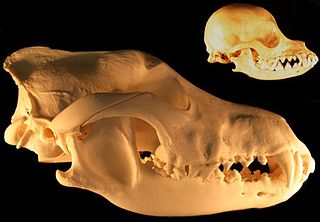
Domestication syndrome refers to two sets of phenotypic traits that are common to either domesticated plants or domesticated animals.
This glossary of genetics and evolutionary biology is a list of definitions of terms and concepts used in the study of genetics and evolutionary biology, as well as sub-disciplines and related fields, with an emphasis on classical genetics, quantitative genetics, population biology, phylogenetics, speciation, and systematics. Overlapping and related terms can be found in Glossary of cellular and molecular biology, Glossary of ecology, and Glossary of biology.
Eukaryote hybrid genomes result from interspecific hybridization, where closely related species mate and produce offspring with admixed genomes. The advent of large-scale genomic sequencing has shown that hybridization is common, and that it may represent an important source of novel variation. Although most interspecific hybrids are sterile or less fit than their parents, some may survive and reproduce, enabling the transfer of adaptive variants across the species boundary, and even result in the formation of novel evolutionary lineages. There are two main variants of hybrid species genomes: allopolyploid, which have one full chromosome set from each parent species, and homoploid, which are a mosaic of the parent species genomes with no increase in chromosome number.
In biology, parallel speciation is a type of speciation where there is repeated evolution of reproductively isolating traits via the same mechanisms occurring between separate yet closely related species inhabiting different environments. This leads to a circumstance where independently evolved lineages have developed reproductive isolation from their ancestral lineage, but not from other independent lineages that inhabit similar environments. In order for parallel speciation to be confirmed, there is a set of three requirements that has been established that must be met: there must be phylogenetic independence between the separate populations inhabiting similar environments to ensure that the traits responsible for reproductive isolation evolved separately, there must be reproductive isolation not only between the ancestral population and the descendent population, but also between descendent populations that inhabit dissimilar environments, and descendent populations that inhabit similar environments must not be reproductively isolated from one another. To determine if natural selection specifically is the cause of parallel speciation, a fourth requirement has been established that includes identifying and testing an adaptive mechanism, which eliminates the possibility of a genetic factor such as polyploidy being the responsible agent.
References
- 1 2 Bellucci, E (2007). Islands of domestication in the common bean (Phaseolus vulgaris L.) genome. Proceedings of the 51st Italian Society of Agricultural Genetics Annual Congress, Riva del Garda, Italy. ISBN 978-88-900622-7-8.
- ↑ Turner, T.L. (2005). "Genomic Islands of Speciation in Anopheles gambiae". PLOS Biology. 3 (9): e285. doi: 10.1371/journal.pbio.0030285 . PMC 1182689 . PMID 16076241.
- ↑ Noor M.A.F. (2009). "Islands of Speciation or Mirages in the Desert? Examining the Role of Restricted Recombination in Maintaining Species". Heredity (Edinb). 103 (6): 439–444. doi:10.1038/hdy.2009.151. PMC 2809014 . PMID 19920849.
- ↑ Cruikshank, T.E. (2014). "Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow". Molecular Ecology. 23 (13): 3133–3157. Bibcode:2014MolEc..23.3133C. doi: 10.1111/mec.12796 . PMID 24845075.
- ↑ Frantz, L (2015). "Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes". Nat. Genet. 47 (10): 1141–8. doi:10.1038/ng.3394. PMID 26323058. S2CID 205350534.