Related Research Articles

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in numerous biological pathways, including digestion of ingested proteins, protein catabolism, and cell signaling.

Serine proteases are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like.

Secretases are enzymes that "snip" pieces off a longer protein that is embedded in the cell membrane.

Beta-secretase 1, also known as beta-site amyloid precursor protein cleaving enzyme 1, beta-site APP cleaving enzyme 1 (BACE1), membrane-associated aspartic protease 2, memapsin-2, aspartyl protease 2, and ASP2, is an enzyme that in humans is encoded by the BACE1 gene. Expression of BACE1 is observed mainly in neurons.
In molecular biology, the Signal Peptide Peptidase (SPP) is a type of protein that specifically cleaves parts of other proteins. It is an intramembrane aspartyl protease with the conserved active site motifs 'YD' and 'GxGD' in adjacent transmembrane domains (TMDs). Its sequences is highly conserved in different vertebrate species. SPP cleaves remnant signal peptides left behind in membrane by the action of signal peptidase and also plays key roles in immune surveillance and the maturation of certain viral proteins.
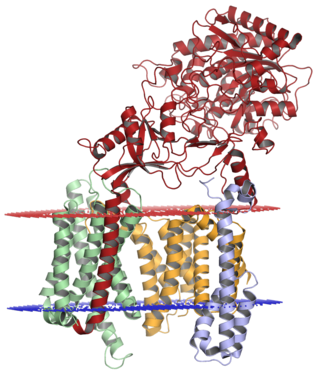
Gamma secretase is a multi-subunit protease complex, itself an integral membrane protein, that cleaves single-pass transmembrane proteins at residues within the transmembrane domain. Proteases of this type are known as intramembrane proteases. The most well-known substrate of gamma secretase is amyloid precursor protein, a large integral membrane protein that, when cleaved by both gamma and beta secretase, produces a short 37-43 amino acid peptide called amyloid beta whose abnormally folded fibrillar form is the primary component of amyloid plaques found in the brains of Alzheimer's disease patients. Gamma secretase is also critical in the related processing of several other type I integral membrane proteins, such as Notch, ErbB4, E-cadherin, N-cadherin, ephrin-B2, or CD44.

Presenilins are a family of related multi-pass transmembrane proteins which constitute the catalytic subunits of the gamma-secretase intramembrane protease protein complex. They were first identified in screens for mutations causing early onset forms of familial Alzheimer's disease by Peter St George-Hyslop. Vertebrates have two presenilin genes, called PSEN1 that codes for presenilin 1 (PS-1) and PSEN2 that codes for presenilin 2 (PS-2). Both genes show conservation between species, with little difference between rat and human presenilins. The nematode worm C. elegans has two genes that resemble the presenilins and appear to be functionally similar, sel-12 and hop-1.
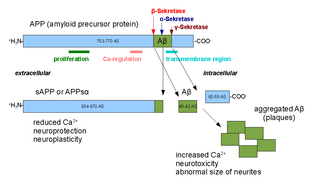
Alpha secretases are a family of proteolytic enzymes that cleave amyloid precursor protein (APP) in its transmembrane region. Specifically, alpha secretases cleave within the fragment that gives rise to the Alzheimer's disease-associated peptide amyloid beta when APP is instead processed by beta secretase and gamma secretase. The alpha-secretase pathway is the predominant APP processing pathway. Thus, alpha-secretase cleavage precludes amyloid beta formation and is considered to be part of the non-amyloidogenic pathway in APP processing. Alpha secretases are members of the ADAM family, which are expressed on the surfaces of cells and anchored in the cell membrane. Several such proteins, notably ADAM10, have been identified as possessing alpha-secretase activity. Upon cleavage by alpha secretases, APP releases its extracellular domain - a fragment known as APPsα - into the extracellular environment in a process known as ectodomain shedding.

Nicastrin, also known as NCSTN, is a protein that in humans is encoded by the NCSTN gene.
APH-1 is a protein gene product originally identified in the Notch signaling pathway in Caenorhabditis elegans as a regulator of the cell-surface localization of nicastrin. APH-1 homologs in other organisms, including humans, have since been identified as components of the gamma secretase complex along with the catalytic subunit presenilin and the regulatory subunits nicastrin and PEN-2. The gamma-secretase complex is a multimeric protease responsible for the intramembrane proteolysis of transmembrane proteins such as the Notch protein and amyloid precursor protein (APP). Gamma-secretase cleavage of APP is one of two proteolytic steps required to generate the peptide known as amyloid beta, whose misfolded form is implicated in the causation of Alzheimer's disease. All of the components of the gamma-secretase complex undergo extensive post-translational modification, especially proteolytic activation; APH-1 and PEN-2 are regarded as regulators of the maturation process of the catalytic component presenilin. APH-1 contains a conserved alpha helix interaction motif glycine-X-X-X-glycine (GXXXG) that is essential to both assembly of the gamma secretase complex and to the maturation of the components.

PSENEN, formally PEN-2, is a protein that is a regulatory component of the gamma secretase complex, a protease complex responsible for proteolysis of transmembrane proteins such as the Notch protein and amyloid precursor protein (APP). The gamma secretase complex consists of PEN-2, APH-1, nicastrin, and the catalytic subunit presenilin. PEN-2 is a 101-amino acid integral membrane protein likely with a topology such that both the N-terminus and the C-terminus face first the lumen of the endoplasmic reticulum and later the extracellular environment. Biochemical studies have shown that a conserved sequence motif D-Y-L-S-F at the C-terminus, as well as the overall length of the C-terminal tail, is required for the formation of an active gamma secretase complex.

Presenilin-1(PS-1) is a presenilin protein that in humans is encoded by the PSEN1 gene. Presenilin-1 is one of the four core proteins in the gamma secretase complex, which is considered to play an important role in generation of amyloid beta (Aβ) from amyloid-beta precursor protein (APP). Accumulation of amyloid beta is associated with the onset of Alzheimer's disease.

Presenilin-2 is a protein that is encoded by the PSEN2 gene.
Membrane-bound transcription factor site-2 protease, also known as S2P endopeptidase or site-2 protease (S2P), is an enzyme encoded by the MBTPS2 gene which liberates the N-terminal fragment of sterol regulatory element-binding protein (SREBP) transcription factors from membranes. S2P cleaves the transmembrane domain of SREPB, making it a member of the class of intramembrane proteases.

Minor histocompatibility antigen H13 is a protein that in humans is encoded by the HM13 gene.

Signal peptide peptidase-like 2B, also known as SPPL2B, is a human gene.

Signal peptide peptidase-like 2A, also known as SPPL2A, is a human gene.

Presenilins-associated rhomboid-like protein, mitochondrial (PSARL), also known as PINK1/PGAM5-associated rhomboid-like protease (PARL), is an inner mitochondrial membrane protein that in humans is encoded by the PARL gene on chromosome 3. It is a member of the rhomboid family of intramembrane serine proteases. This protein is involved in signal transduction and apoptosis, as well as neurodegenerative diseases and type 2 diabetes.

The rhomboid proteases are a family of enzymes that exist in almost all species. They are proteases: they cut the polypeptide chain of other proteins. This proteolytic cleavage is irreversible in cells, and an important type of cellular regulation. Although proteases are one of the earliest and best studied class of enzyme, rhomboids belong to a much more recently discovered type: the intramembrane proteases. What is unique about intramembrane proteases is that their active sites are buried in the lipid bilayer of cell membranes, and they cleave other transmembrane proteins within their transmembrane domains. About 30% of all proteins have transmembrane domains, and their regulated processing often has major biological consequences. Accordingly, rhomboids regulate many important cellular processes, and may be involved in a wide range of human diseases.

Rhomboid-related protein 2 is a protein that in humans is encoded by the RHBDL2 gene.
References
- 1 2 Brown, MS; Ye, J; Rawson, RB; Goldstein, JL (18 February 2000). "Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans". Cell. 100 (4): 391–8. doi: 10.1016/S0092-8674(00)80675-3 . PMID 10693756.
- ↑ Urban, S; Freeman, M (October 2002). "Intramembrane proteolysis controls diverse signalling pathways throughout evolution". Current Opinion in Genetics & Development. 12 (5): 512–8. doi:10.1016/s0959-437x(02)00334-9. PMID 12200155.
- ↑ Wolfe, MS; Kopan, R (20 August 2004). "Intramembrane proteolysis: theme and variations". Science. 305 (5687): 1119–23. doi:10.1126/science.1096187. PMID 15326347.
- ↑ Erez, E; Fass, D; Bibi, E (21 May 2009). "How intramembrane proteases bury hydrolytic reactions in the membrane". Nature. 459 (7245): 371–8. doi:10.1038/nature08146. PMID 19458713.
- 1 2 3 4 5 6 Kühnle, Nathalie; Dederer, Verena; Lemberg, Marius K. (15 August 2019). "Intramembrane proteolysis at a glance: from signalling to protein degradation". Journal of Cell Science. 132 (16): jcs217745. doi: 10.1242/jcs.217745 .
- ↑ Koonin, EV; Makarova, KS; Rogozin, IB; Davidovic, L; Letellier, MC; Pellegrini, L (2003). "The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers". Genome Biology. 4 (3): R19. doi: 10.1186/gb-2003-4-3-r19 . PMC 153459 . PMID 12620104.
- ↑ Lemberg, M. K.; Freeman, M. (1 November 2007). "Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases". Genome Research. 17 (11): 1634–1646. doi:10.1101/gr.6425307. PMC 2045146 . PMID 17938163.
- ↑ Wolfe, M. S. (3 February 2009). "Intramembrane-cleaving Proteases". Journal of Biological Chemistry. 284 (21): 13969–13973. doi: 10.1074/jbc.R800039200 . PMC 2682844 . PMID 19189971.
- 1 2 Rawson, RB; Zelenski, NG; Nijhawan, D; Ye, J; Sakai, J; Hasan, MT; Chang, TY; Brown, MS; Goldstein, JL (December 1997). "Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs". Molecular Cell. 1 (1): 47–57. doi: 10.1016/s1097-2765(00)80006-4 . PMID 9659902.
- ↑ Wolfe, MS; Xia, W; Ostaszewski, BL; Diehl, TS; Kimberly, WT; Selkoe, DJ (8 April 1999). "Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity". Nature. 398 (6727): 513–7. doi:10.1038/19077. PMID 10206644.
- ↑ De Strooper, B; Annaert, W; Cupers, P; Saftig, P; Craessaerts, K; Mumm, JS; Schroeter, EH; Schrijvers, V; Wolfe, MS; Ray, WJ; Goate, A; Kopan, R (8 April 1999). "A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain". Nature. 398 (6727): 518–22. doi:10.1038/19083. PMID 10206645.
- ↑ Weihofen, A; Binns, K; Lemberg, MK; Ashman, K; Martoglio, B (21 June 2002). "Identification of signal peptide peptidase, a presenilin-type aspartic protease". Science. 296 (5576): 2215–8. doi:10.1126/science.1070925. PMID 12077416.
- ↑ Friedmann, E; Hauben, E; Maylandt, K; Schleeger, S; Vreugde, S; Lichtenthaler, SF; Kuhn, PH; Stauffer, D; Rovelli, G; Martoglio, B (August 2006). "SPPL2a and SPPL2b promote intramembrane proteolysis of TNFalpha in activated dendritic cells to trigger IL-12 production". Nature Cell Biology. 8 (8): 843–8. doi:10.1038/ncb1440. PMID 16829952.
- ↑ Urban, S; Lee, JR; Freeman, M (19 October 2001). "Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases". Cell. 107 (2): 173–82. doi: 10.1016/s0092-8674(01)00525-6 . PMID 11672525.
- ↑ Hampton, Shahienaz E.; Dore, Timothy M.; Schmidt, Walter K. (4 March 2018). "Rce1: mechanism and inhibition". Critical Reviews in Biochemistry and Molecular Biology. 53 (2): 157–174. doi: 10.1080/10409238.2018.1431606 . PMC 5874806 . PMID 29424242.
- ↑ Manolaridis, Ioannis; Kulkarni, Kiran; Dodd, Roger B.; Ogasawara, Satoshi; Zhang, Ziguo; Bineva, Ganka; O’Reilly, Nicola; Hanrahan, Sarah J.; Thompson, Andrew J.; Cronin, Nora; Iwata, So; Barford, David (December 2013). "Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1". Nature. 504 (7479): 301–305. doi:10.1038/nature12754. PMC 3864837 . PMID 24291792.
- 1 2 3 4 Sun, Linfeng; Li, Xiaochun; Shi, Yigong (April 2016). "Structural biology of intramembrane proteases: mechanistic insights from rhomboid and S2P to γ-secretase". Current Opinion in Structural Biology. 37: 97–107. doi:10.1016/j.sbi.2015.12.008.
- 1 2 3 4 Beard, Hester A.; Barniol-Xicota, Marta; Yang, Jian; Verhelst, Steven H. L. (15 November 2019). "Discovery of Cellular Roles of Intramembrane Proteases". ACS Chemical Biology. 14 (11): 2372–2388. doi:10.1021/acschembio.9b00404.
- 1 2 Sanders, Charles R; Hutchison, James M (August 2018). "Membrane properties that shape the evolution of membrane enzymes". Current Opinion in Structural Biology. 51: 80–91. doi:10.1016/j.sbi.2018.03.013. PMC 6158105 . PMID 29597094.
- ↑ Güner G, Lichtenthaler SF (September 2020). "The substrate repertoire of γ-secretase/presenilin". Seminars in Cell & Developmental Biology. 105: 27–42. doi: 10.1016/j.semcdb.2020.05.019 . PMID 32616437.
- 1 2 Paschkowsky, Sandra; Hsiao, Jacqueline Melissa; Young, Jason C.; Munter, Lisa Marie (June 2019). "The discovery of proteases and intramembrane proteolysis". Biochemistry and Cell Biology. 97 (3): 265–269. doi:10.1139/bcb-2018-0186.
- ↑ Selkoe, Dennis J. (August 1996). "Amyloid β-Protein and the Genetics of Alzheimer's Disease". Journal of Biological Chemistry. 271 (31): 18295–18298. doi: 10.1074/jbc.271.31.18295 .