Related Research Articles

Poliovirus, the causative agent of polio, is a serotype of the species Enterovirus C, in the family of Picornaviridae. There are three poliovirus serotypes: types 1, 2, and 3.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.

Proteinase 3, also known as PRTN3, is an enzyme that in humans is encoded by the PRTN3 gene.

Cathepsin S is a protein that in humans is encoded by the CTSS gene. Transcript variants utilizing alternative polyadenylation signals exist for this gene.
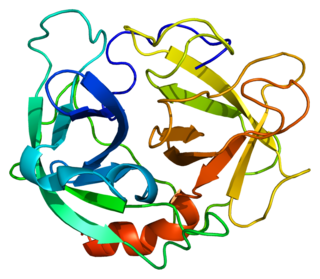
Neutrophil elastase is a serine proteinase in the same family as chymotrypsin and has broad substrate specificity. Neutrophil elastase is secreted by neutrophils during inflammation, and destroys bacteria and host tissue. It also localizes to neutrophil extracellular traps (NETs), via its high affinity for DNA, an unusual property for serine proteases.

TEV protease is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV). It is a member of the PA clan of chymotrypsin-like proteases. Due to its high sequence specificity, TEV protease is frequently used for the controlled cleavage of fusion proteins in vitro and in vivo.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.

HIV-1 protease (PR) is a retroviral aspartyl protease (retropepsin), an enzyme involved with peptide bond hydrolysis in retroviruses, that is essential for the life-cycle of HIV, the retrovirus that causes AIDS. HIV protease cleaves newly synthesized polyproteins at nine cleavage sites to create the mature protein components of an HIV virion, the infectious form of a virus outside of the host cell. Without effective HIV protease, HIV virions remain uninfectious.

Protease activated receptor 2 (PAR2) also known as coagulation factor II (thrombin) receptor-like 1 (F2RL1) or G-protein coupled receptor 11 (GPR11) is a protein that in humans is encoded by the F2RL1 gene. PAR2 modulates inflammatory responses, obesity, metabolism, cancers and acts as a sensor for proteolytic enzymes generated during infection. In humans, we can find PAR2 in the stratum granulosum layer of epidermal keratinocytes. Functional PAR2 is also expressed by several immune cells such as eosinophils, neutrophils, monocytes, macrophages, dendritic cells, mast cells and T cells.
Kexin is a prohormone-processing protease, specifically a yeast serine peptidase, found in the budding yeast. It catalyzes the cleavage of -Lys-Arg- and -Arg-Arg- bonds to process yeast alpha-factor pheromone and killer toxin precursors. The human homolog is PCSK4. It is a family of subtilisin-like peptidases. Even though there are a few prokaryote kexin-like peptidases, all kexins are eukaryotes. The enzyme is encoded by the yeast gene KEX2, and usually referred to in the scientific community as Kex2p. It shares structural similarities with the bacterial protease subtilisin. The first mammalian homologue of this protein to be identified was furin. In the mammal, kexin-like peptidases function in creating and regulating many differing proproteins.
Eckard Wimmer is a German American virologist, organic chemist and distinguished professor of molecular genetics and microbiology at Stony Brook University. He is best known for his seminal work on the molecular biology of poliovirus and the first chemical synthesis of a viral genome capable of infection and subsequent production of live viruses.
Streptogrisin A is an enzyme. This enzyme catalyses the following chemical reaction
Togavirin is an enzyme. This enzyme catalyses the following chemical reaction
Pestivirus NS3 polyprotein peptidase is an enzyme. This enzyme catalyses the following chemical reaction

Picornain 3C is a protease found in picornaviruses, which cleaves peptide bonds of non-terminal sequences. Picornain 3C’s endopeptidase activity is primarily responsible for the catalytic process of selectively cleaving Gln-Gly bonds in the polyprotein of poliovirus and with substitution of Glu for Gln, and Ser or Thr for Gly in other picornaviruses. Picornain 3C are cysteine proteases related by amino acid sequence to trypsin-like serine proteases. Picornain 3C is encoded by enteroviruses, rhinoviruses, aphtoviruses and cardioviruses. These genera of picoviruses cause a wide range of infections in humans and mammals.

The 3C-like protease (3CLpro) or main protease (Mpro), formally known as C30 endopeptidase or 3-chymotrypsin-like protease, is the main protease found in coronaviruses. It cleaves the coronavirus polyprotein at eleven conserved sites. It is a cysteine protease and a member of the PA clan of proteases. It has a cysteine-histidine catalytic dyad at its active site and cleaves a Gln–(Ser/Ala/Gly) peptide bond.
Bacillolysin is an enzyme. This enzyme catalyses the following chemical reaction
A protein superfamily is the largest grouping (clade) of proteins for which common ancestry can be inferred. Usually this common ancestry is inferred from structural alignment and mechanistic similarity, even if no sequence similarity is evident. Sequence homology can then be deduced even if not apparent. Superfamilies typically contain several protein families which show sequence similarity within each family. The term protein clan is commonly used for protease and glycosyl hydrolases superfamilies based on the MEROPS and CAZy classification systems.

The PA clan is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have identity of <10%. The clan contains both cysteine and serine proteases. PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.
References
- ↑ Bazan JF, Fletterick RJ (November 1988). "Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications". Proceedings of the National Academy of Sciences of the United States of America. 85 (21): 7872–6. Bibcode:1988PNAS...85.7872B. doi: 10.1073/pnas.85.21.7872 . PMC 282299 . PMID 3186696.
- ↑ König H, Rosenwirth B (April 1988). "Purification and partial characterization of poliovirus protease 2A by means of a functional assay". Journal of Virology. 62 (4): 1243–50. doi:10.1128/JVI.62.4.1243-1250.1988. PMC 253133 . PMID 2831385.
- ↑ Kräusslich HG, Wimmer E (1988). "Viral proteinases". Annual Review of Biochemistry. 57: 701–54. doi:10.1146/annurev.bi.57.070188.003413. PMID 3052288.