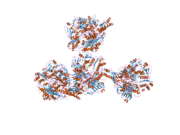Uric acid is a heterocyclic compound of carbon, nitrogen, oxygen, and hydrogen with the formula C5H4N4O3. It forms ions and salts known as urates and acid urates, such as ammonium acid urate. Uric acid is a product of the metabolic breakdown of purine nucleotides, and it is a normal component of urine. High blood concentrations of uric acid can lead to gout and are associated with other medical conditions, including diabetes and the formation of ammonium acid urate kidney stones.
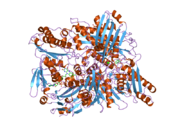
Xanthine oxidase is a form of xanthine oxidoreductase, a type of enzyme that generates reactive oxygen species. These enzymes catalyze the oxidation of hypoxanthine to xanthine and can further catalyze the oxidation of xanthine to uric acid. These enzymes play an important role in the catabolism of purines in some species, including humans.

Xanthinuria, also known as xanthine oxidase deficiency, is a rare genetic disorder causing the accumulation of xanthine. It is caused by a deficiency of the enzyme xanthine oxidase.
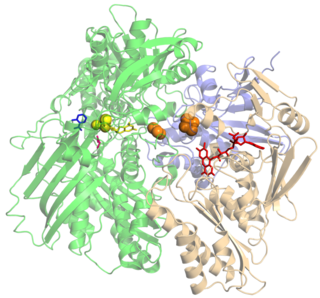
L-Gulonolactone oxidase is an enzyme that produces vitamin C. It is expressed in most mammals, but is non-functional in Haplorrhini, in some bats, and in guinea pigs. It catalyzes the reaction of L-gulono-1,4-lactone with oxygen to form L-xylo-hex-3-gulonolactone (2-keto-gulono-γ-lactone) and hydrogen peroxide. It uses FAD as a cofactor. The L-xylo-hex-3-gulonolactone then converts to ascorbic acid spontaneously, without enzymatic action. The structure of L-gulonolactone oxidase in rats helps identify characteristics of this enzyme.

Dihydrolipoamide dehydrogenase (DLD), also known as dihydrolipoyl dehydrogenase, mitochondrial, is an enzyme that in humans is encoded by the DLD gene. DLD is a flavoprotein enzyme that oxidizes dihydrolipoamide to lipoamide.

Fatty aldehyde dehydrogenase is an aldehyde dehydrogenase enzyme that in human is encoded in the ALDH3A2 gene on chromosome 17. Aldehyde dehydrogenase enzymes function to remove toxic aldehydes that are generated by the metabolism of alcohol and by lipid peroxidation.

UDP-glucose 6-dehydrogenase is a cytosolic enzyme that in humans is encoded by the UGDH gene.
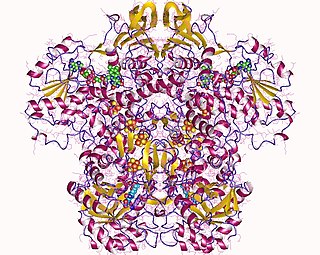
In enzymology, a dihydropyrimidine dehydrogenase (NADP+) (EC 1.3.1.2) is an enzyme that catalyzes the chemical reaction
In enzymology, an aldehyde dehydrogenase (FAD-independent) (EC 1.2.99.7) is an enzyme that catalyzes the chemical reaction
In enzymology, a phenylalanine dehydrogenase (EC 1.4.1.20) is an enzyme that catalyzes the chemical reaction

Aldo-keto reductase family 1 member C1 also known as 20α-hydroxysteroid dehydrogenase, 3α-hydroxysteroid dehydrogenase, and dihydrodiol dehydrogenase 1/2 is an enzyme that in humans is encoded by the AKR1C1 gene.

D-bifunctional protein (DBP), also known as peroxisomal multifunctional enzyme type 2 (MFP-2), as well as 17β-hydroxysteroid dehydrogenase type IV is a protein that in humans is encoded by the HSD17B4 gene. It's an alcohol oxidoreductase, specifically 17β-Hydroxysteroid dehydrogenase. It is involved in fatty acid β-oxidation and steroid metabolism.

NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial (NDUFV2) is an enzyme that in humans is encoded by the NDUFV2 gene. The encoded protein, NDUFV2, is a subunit of complex I of the mitochondrial respiratory chain, which is located on the inner mitochondrial membrane and involved in oxidative phosphorylation. Mutations in this gene are implicated in Parkinson's disease, bipolar disorder, schizophrenia, and have been found in one case of early onset hypertrophic cardiomyopathy and encephalopathy.

NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 is an enzyme that in humans is encoded by the NDUFB9 gene. NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 9 is an accessory subunit of the NADH dehydrogenase (ubiquinone) complex, located in the mitochondrial inner membrane. It is also known as Complex I and is the largest of the five complexes of the electron transport chain.

Molybdenum cofactor sulfurase is an enzyme that in humans is encoded by the MOCOS gene.

NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial is an enzyme that in humans is encoded by the NDUFB11 gene. NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 11 is an accessory subunit of the NADH dehydrogenase (ubiquinone) complex, located in the mitochondrial inner membrane. It is also known as Complex I and is the largest of the five complexes of the electron transport chain. NDUFB11 mutations have been associated with linear skin defects with multiple congenital anomalies 3 and mitochondrial complex I deficiency.

Aldehyde oxidase 1 is an enzyme that in humans is encoded by the AOX1 gene.

Short-chain acyl-CoA dehydrogenase is an enzyme with systematic name short-chain acyl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase. This enzyme catalyses the following chemical reaction
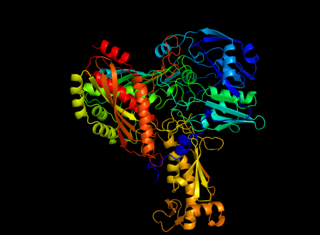
Caffeine dehydrogenase, commonly referred to in scientific literature as caffeine oxidase, is an enzyme with the systematic name caffeine:ubiquinone oxidoreductase. The enzyme is most well known for its ability to directly oxidize caffeine, a type of methylxanthine, to trimethyluric acid. Caffeine dehydrogenase can be found in bacterium Pseudomonas sp. CBB1 and in several species within the genera Alcaligenes, Rhodococcus, and Klebsiella.

NDUFA4, mitochondrial complex associated is a protein that in humans is encoded by the NDUFA4 gene. The NDUFA4 protein was first described to be a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. However, recent research has described NDUFA4 as a subunit of cytochrome c oxidase. Mutations in the NDUFA4 gene are associated with Leigh's syndrome.