External links
- Basic+helix-loop-helix+leucine+zipper+transcription+factors at the US National Library of Medicine Medical Subject Headings (MeSH)
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| This genetics article is a stub. You can help Wikipedia by expanding it. |
Basic helix-loop-helix leucine zipper transcription factors are, as their name indicates, transcription factors containing both Basic helix-loop-helix and leucine zipper motifs.
Examples include Microphthalmia-associated transcription factor and Sterol regulatory element binding protein (SREBP).
| This genetics article is a stub. You can help Wikipedia by expanding it. |

DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others.
Microphthalmia, also referred as microphthalmos, is a developmental disorder of the eye in which one or both eyes are abnormally small and have anatomic malformations. It is different from nanophthalmos in which the eye is small in size but has no anatomical alterations.

A leucine zipper is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization.
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure.

Sterol regulatory element-binding proteins (SREBPs) are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors. Unactivated SREBPs are attached to the nuclear envelope and endoplasmic reticulum membranes. In cells with low levels of sterols, SREBPs are cleaved to a water-soluble N-terminal domain that is translocated to the nucleus. These activated SREBPs then bind to specific sterol regulatory element DNA sequences, thus upregulating the synthesis of enzymes involved in sterol biosynthesis. Sterols in turn inhibit the cleavage of SREBPs and therefore synthesis of additional sterols is reduced through a negative feed back loop.

CCAAT-enhancer-binding proteins is a family of transcription factors composed of six members, named from C/EBPα to C/EBPζ. They promote the expression of certain genes through interaction with their promoters. Once bound to DNA, C/EBPs can recruit so-called co-activators that in turn can open up chromatin structure or recruit basal transcription factors.
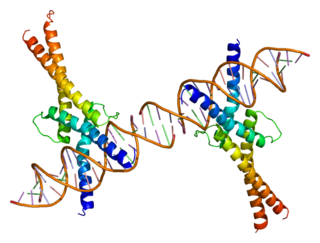
Sterol regulatory element-binding transcription factor 1 (SREBF1) also known as sterol regulatory element-binding protein 1 (SREBP-1) is a protein that in humans is encoded by the SREBF1 gene.

Sterol regulatory element-binding protein 2 (SREBP-2) also known as sterol regulatory element binding transcription factor 2 (SREBF2) is a protein that in humans is encoded by the SREBF2 gene.

Upstream stimulatory factor 1 is a protein that in humans is encoded by the USF1 gene.

MAX is a gene that in humans encodes the MAX transcription factor.

Upstream stimulatory factor 2 is a protein that in humans is encoded by the USF2 gene.

Transcription factor E3 is a protein that in humans is encoded by the TFE3 gene.

Hepatic leukemia factor is a protein that in humans is encoded by the HLF gene.

Neural retina-specific leucine zipper protein is a protein that in humans is encoded by the NRL gene.

Max-like protein X is a protein that in humans is encoded by the MLX gene.

Max-interacting transcriptional repressor MAD4 is a protein that in humans is encoded by the MXD4 gene.
Chimeric nucleases are an example of engineered proteins which must comprise a DNA-binding domain to give sequence specificity and a nuclease domain for DNA cleavage.

The Basic Leucine Zipper Domain is found in many DNA binding eukaryotic proteins. One part of the domain contains a region that mediates sequence specific DNA binding properties and the leucine zipper that is required to hold together (dimerize) two DNA binding regions. The DNA binding region comprises a number of basic amino acids such as arginine and lysine. Proteins containing this domain are transcription factors.
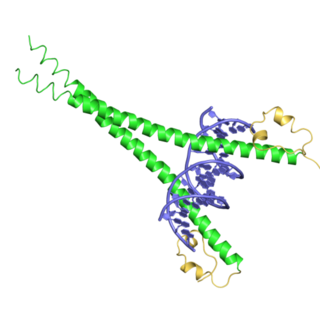
bZIP Maf is a domain found in Maf transcription factor proteins. It contains a leucine zipper (bZIP) domain, which mediates the transcription factor's dimerization and DNA binding properties. The Maf extended homology region (EHR) is present at the N-terminus of the protein. This region exists only within the Maf family and allows the family to recognize longer DNA motifs than other leucine zippers. These motifs are termed the Maf recognition element (MARE) and is 13 or 14 base pairs long. In particular, the two residues at the beginning of helix H2 are positioned to recognise the flanking region of the DNA. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF2-E2 transcription factor.
A microprotein (miP) is a small protein encoded from small open reading frame (smORF). They are a class of protein with a single protein domain that are related to multidomain proteins. Microproteins regulate larger multidomain proteins at the post-translational level. Microproteins are analogous to microRNAs (miRNAs) and heterodimerize with their targets causing dominant and negative effects. In animals and plants, microproteins have been found to greatly influence the biological processes. Because of microproteins' dominant effects on their targets, microproteins are currently being studied for potential applications in biotechnology.