Related Research Articles

Caenorhabditis elegans is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a blend of the Greek caeno- (recent), rhabditis (rod-like) and Latin elegans (elegant). In 1900, Maupas initially named it Rhabditides elegans. Osche placed it in the subgenus Caenorhabditis in 1952, and in 1955, Dougherty raised Caenorhabditis to the status of genus.

A microRNA is a small non-coding RNA molecule found in plants, animals and some viruses, that functions in RNA silencing and post-transcriptional regulation of gene expression. miRNAs function via base-pairing with complementary sequences within mRNA molecules. As a result, these mRNA molecules are silenced, by one or more of the following processes: (1) Cleavage of the mRNA strand into two pieces, (2) Destabilization of the mRNA through shortening of its poly(A) tail, and (3) Less efficient translation of the mRNA into proteins by ribosomes.

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as transfer RNA (tRNA) or small nuclear RNA (snRNA) genes, the product is a functional RNA. Gene expression is summarized in the Central Dogma first formulated by Francis Crick in 1958, further developed in his 1970 article, and expanded by the subsequent discoveries of reverse transcription and RNA replication.

In molecular genetics, the three prime untranslated region (3'-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3'-UTR often contains regulatory regions that post-transcriptionally influence gene expression.

Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.
An asymmetric cell division produces two daughter cells with different cellular fates. This is in contrast to symmetric cell divisions which give rise to daughter cells of equivalent fates. Notably, stem cells divide asymmetrically to give rise to two distinct daughter cells: one copy of the original stem cell as well as a second daughter programmed to differentiate into a non-stem cell fate.
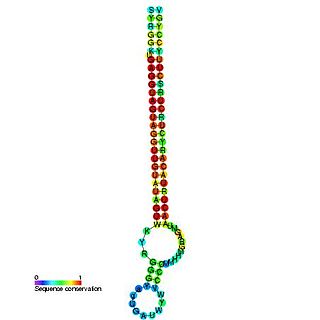
The Let-7 microRNA precursor was identified from a study of developmental timing in C. elegans, and was later shown to be part of a much larger class of non-coding RNAs termed microRNAs. miR-98 microRNA precursor from human is a let-7 family member. Let-7 miRNAs have now been predicted or experimentally confirmed in a wide range of species (MIPF0000002). miRNAs are initially transcribed in long transcripts called primary miRNAs (pri-miRNAs), which are processed in the nucleus by Drosha and Pasha to hairpin structures of about 70 nucleotide. These precursors (pre-miRNAs) are exported to the cytoplasm by exportin5, where they are subsequently processed by the enzyme Dicer to a ~22 nucleotide mature miRNA. The involvement of Dicer in miRNA processing demonstrates a relationship with the phenomenon of RNA interference.

In molecular biology lin-4 is a microRNA (miRNA) that was identified from a study of developmental timing in the nematode Caenorhabditis elegans. It was the first to be discovered of the miRNAs, a class of non-coding RNAs involved in gene regulation. miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a 21 nucleotide product. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complete or partial complementarity to mRNA. The lin-4 gene has been found to lie within a 4.11kb intron of a separate host gene.

The miR-8 microRNA precursor, is a short non-coding RNA gene involved in gene regulation. miR-8 in Drosophila melanogaster is expressed from the 3' arm of related precursor hairpins, along with miR-200, miR-236, miR-429 and human and mouse homolog miR-141. Members of this precursor family have now been predicted or experimentally confirmed in a wide range of species. The bounds of the precursors are predicted based on conservation and base pairing and are not generally known.

The miR-10 microRNA precursor is a short non-coding RNA gene involved in gene regulation. It is part of an RNA gene family which contains miR-10, miR-51, miR-57, miR-99 and miR-100. miR-10, miR-99 and miR-100 have now been predicted or experimentally confirmed in a wide range of species. mir-51 and mir-57 have currently only been identified in the nematode Caenorhabditis elegans.
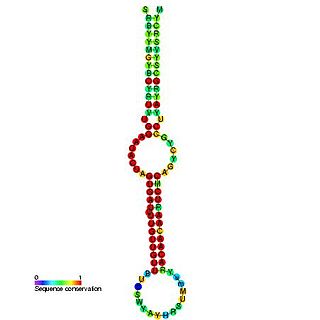
This family represents the microRNA (miRNA) precursor mir-7. This miRNA has been predicted or experimentally confirmed in a wide range of species. miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 5' arm of the precursor. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The involvement of Dicer in miRNA processing suggests a relationship with the phenomenon of RNA interference.

The miR-92 microRNAs are short single stranded non-protein coding RNA fragments initially discovered incorporated into an RNP complex with a proposed role of processing RNA molecules and further RNP assembly. Mir-92 has been mapped to the human genome as part of a larger cluster at chromosome 13q31.3, where it is 22 nucleotides in length but exists in the genome as part of a longer precursor sequence. There is an exact replica of the mir-92 precursor on the X chromosome. MicroRNAs are endogenous triggers of the RNAi pathway which involves several ribonucleic proteins (RNPs) dedicated to repressing mRNA molecules via translation inhibition and/or induction of mRNA cleavage. miRNAs are themselves matured from their long RNA precursors by ribonucleic proteins as part of a 2 step biogenesis mechanism involving RNA polymerase 2.
Gary Bruce Ruvkun is an American molecular biologist at Massachusetts General Hospital and professor of genetics at Harvard Medical School in Boston. Ruvkun discovered the mechanism by which lin-4, the first microRNA (miRNA) discovered by Victor Ambros, regulates the translation of target messenger RNAs via imperfect base-pairing to those targets, and discovered the second miRNA, let-7, and that it is conserved across animal phylogeny, including in humans. These miRNA discoveries revealed a new world of RNA regulation at an unprecedented small size scale, and the mechanism of that regulation. Ruvkun also discovered many features of insulin-like signaling in the regulation of aging and metabolism. He was elected a Member of the American Philosophical Society in 2019.

In molecular biology miR-132 microRNA is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms, generally reducing protein levels through the cleavage of mRNAs or the repression of their translation. Several targets for miR-132 have been described, including mediators of neurological development, synaptic transmission, inflammation and angiogenesis.

In molecular biology, miR-137 is a short non-coding RNA molecule that functions to regulate the expression levels of other genes by various mechanisms. miR-137 is located on human chromosome 1p22 and has been implicated to act as a tumor suppressor in several cancer types including colorectal cancer, squamous cell carcinoma and melanoma via cell cycle control.

In molecular biology, mir-433 is a short non-coding RNA molecule. MicroRNAs (miRNAs) function as posttranscriptional regulators of expression levels of other genes by several mechanisms. They play roles in development, metabolism and carcinogenesis.
In molecular biology mir-84 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-241 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-939 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
NamiRNAs are a type of miRNAs present in the nucleus, which can activate [[gene expression]] by binding to the enhancer, and therefore were named nuclear activating miRNAs (NamiRNAs), such as miR-24-1 and miR-26. These miRNAs loci are enriched with epigenetic markers that display enhancer activity like histone H3K27ac, P300/CBP, and DNaseI high-sensitivity loci. These NamiRNAs are able to activate the related enhancers and co-work with them to up-regulate the expression of neighboring genes. NamiRNAs are able to promote global gene transcription by binding their targeted enhancers in whole genome level.
References
- 1 2 3 Johnston RJ, Hobert O (December 2003). "A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans". Nature. 426 (6968): 845–849. doi:10.1038/nature02255. PMID 14685240.
- ↑ Johnston RJ, Hobert O (December 2005). "A novel C. elegans zinc finger transcription factor, lsy-2, required for the cell type-specific expression of the lsy-6 microRNA". Development. 132 (24): 5451–5460. doi: 10.1242/dev.02163 . PMID 16291785.
- ↑ Didiano D, Hobert O (July 2008). "Molecular architecture of a miRNA-regulated 3′ UTR". RNA. 14 (7): 1297–1317. doi:10.1261/rna.1082708. PMC 2441980 . PMID 18463285.
- ↑ Chang S, Johnston RJ, Frøkjaer-Jensen C, Lockery S, Hobert O (August 2004). "MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode". Nature. 430 (7001): 785–789. doi:10.1038/nature02752. PMID 15306811.