Related Research Articles

MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then gene silence said mRNA molecules by one or more of the following processes:
- Cleavage of mRNA strand into two pieces,
- Destabilization of mRNA by shortening its poly(A) tail, or
- Translation of mRNA into proteins.
Small RNA (sRNA) are polymeric RNA molecules that are less than 200 nucleotides in length, and are usually non-coding. RNA silencing is often a function of these molecules, with the most common and well-studied example being RNA interference (RNAi), in which endogenously expressed microRNA (miRNA) or exogenously derived small interfering RNA (siRNA) induces the degradation of complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies repeat associated small interfering RNA (rasiRNA). Small RNA "is unable to induce RNAi alone, and to accomplish the task it must form the core of the RNA–protein complex termed the RNA-induced silencing complex (RISC), specifically with Argonaute protein".

Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the DICER1 gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments called small interfering RNA and microRNA, respectively. These fragments are approximately 20–25 base pairs long with a two-base overhang on the 3′-end. Dicer facilitates the activation of the RNA-induced silencing complex (RISC), which is essential for RNA interference. RISC has a catalytic component Argonaute, which is an endonuclease capable of degrading messenger RNA (mRNA).
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).

The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi). Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity, which then leads to mRNA cleavage, translation inhibition, and/or the initiation of mRNA decay.
Piwi-interacting RNA (piRNA) is the largest class of small non-coding RNA molecules expressed in animal cells. piRNAs form RNA-protein complexes through interactions with piwi-subfamily Argonaute proteins. These piRNA complexes are mostly involved in the epigenetic and post-transcriptional silencing of transposable elements and other spurious or repeat-derived transcripts, but can also be involved in the regulation of other genetic elements in germ line cells.
RNA activation (RNAa) is a small RNA-guided and Argonaute (Ago)-dependent gene regulation phenomenon in which promoter-targeted short double-stranded RNAs (dsRNAs) induce target gene expression at the transcriptional/epigenetic level. RNAa was first reported in a 2006 PNAS paper by Li et al. who also coined the term "RNAa" as a contrast to RNA interference (RNAi) to describe such gene activation phenomenon. dsRNAs that trigger RNAa have been termed small activating RNA (saRNA). Since the initial discovery of RNAa in human cells, many other groups have made similar observations in different mammalian species including human, non-human primates, rat and mice, plant and C. elegans, suggesting that RNAa is an evolutionarily conserved mechanism of gene regulation.

MicroRNA (miRNA) precursor miR156 is a family of plant non-coding RNA. This microRNA has now been predicted or experimentally confirmed in a range of plant species. Animal miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. miR156 functions in the induction of flowering by suppressing the transcripts of SQUAMOSA-PROMOTER BINDING LIKE (SPL) transcription factors gene family. It was suggested that the loading into ARGONAUTE1 and ARGONAUTE5 is required for miR156 functionality in Arabidopsis thaliana. In plants the precursor sequences may be longer, and the carpel factory (caf) enzyme appears to be involved in processing. In this case the mature sequence comes from the 5' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complementarity to mRNA.
In molecular biology, mir-160 is a microRNA that has been predicted or experimentally confirmed in a range of plant species including Arabidopsis thaliana and Oryza sativa (rice). miR-160 is predicted to bind complementary sites in the untranslated regions of auxin response factor genes to regulate their expression. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.
Trans-acting siRNA are a class of small interfering RNA (siRNA) that repress gene expression through post-transcriptional gene silencing in land plants. Precursor transcripts from TAS loci are polyadenylated and converted to double-stranded RNA, and are then processed into 21-nucleotide-long RNA duplexes with overhangs. These segments are incorporated into an RNA-induced silencing complex (RISC) and direct the sequence-specific cleavage of target mRNA. Ta-siRNAs are classified as siRNA because they arise from double-stranded RNA (dsRNA).
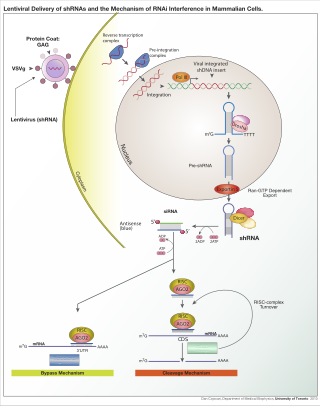
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be developed and find utility as novel therapeutic agents.
In molecular biology mir-383 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-396 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-397 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-408 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-828 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-398 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

DCL1 is a gene in plants that codes for the DCL1 protein, a ribonuclease III enzyme involved in processing double-stranded RNA (dsRNA) and microRNA (miRNA). Although DCL1, also called Endoribonuclease Dicer homolog 1, is named for its homology with the metazoan protein Dicer, its role in miRNA biogenesis is somewhat different, due to substantial differences in miRNA maturation processes between plants and animals, as well due to additional downstream plant-specific pathways, where DCL1 paralogs like DCL4 participate, such Trans-acting siRNA biogenesis.

RNA-directed DNA methylation (RdDM) is a biological process in which non-coding RNA molecules direct the addition of DNA methylation to specific DNA sequences. The RdDM pathway is unique to plants, although other mechanisms of RNA-directed chromatin modification have also been described in fungi and animals. To date, the RdDM pathway is best characterized within angiosperms, and particularly within the model plant Arabidopsis thaliana. However, conserved RdDM pathway components and associated small RNAs (sRNAs) have also been found in other groups of plants, such as gymnosperms and ferns. The RdDM pathway closely resembles other sRNA pathways, particularly the highly conserved RNAi pathway found in fungi, plants, and animals. Both the RdDM and RNAi pathways produce sRNAs and involve conserved Argonaute, Dicer and RNA-dependent RNA polymerase proteins.

James C. Carrington is a plant biologist and the current president of the Donald Danforth Plant Science Center. In 2005 he was elected a fellow of the American Association for the Advancement of Science and in 2008 he was elected to the National Academy of Sciences.
References
- ↑ Mendoza-Soto AB, Sánchez F, Hernández G (2012). "MicroRNAs as regulators in plant metal toxicity response". Frontiers in Plant Science. 3: 105. doi: 10.3389/fpls.2012.00105 . PMC 3356851 . PMID 22661980.
- ↑ Zhang J, Zhang S, Han S, Wu T, Li X, Li W, Qi L (August 2012). "Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis". Planta. 236 (2): 647–57. doi:10.1007/s00425-012-1643-9. PMID 22526500. S2CID 6776890.
- ↑ Jouannet V, Moreno AB, Elmayan T, Vaucheret H, Crespi MD, Maizel A (April 2012). "Cytoplasmic Arabidopsis AGO7 accumulates in membrane-associated siRNA bodies and is required for ta-siRNA biogenesis". The EMBO Journal. 31 (7): 1704–13. doi:10.1038/emboj.2012.20. PMC 3321200 . PMID 22327216.
- ↑ Li F, Orban R, Baker B (June 2012). "SoMART: a web server for plant miRNA, tasiRNA and target gene analysis". The Plant Journal. 70 (5): 891–901. doi:10.1111/j.1365-313X.2012.04922.x. PMID 22268718.
- ↑ Maunoury N, Vaucheret H (2011). Bendahmane M (ed.). "AGO1 and AGO2 act redundantly in miR408-mediated Plantacyanin regulation". PLOS ONE. 6 (12): e28729. Bibcode:2011PLoSO...628729M. doi: 10.1371/journal.pone.0028729 . PMC 3236769 . PMID 22174881.
- ↑ Pérez-Quintero AL, Sablok G, Tatarinova TV, Conesa A, Kuo J, López C (April 2012). "Mining of miRNAs and potential targets from gene oriented clusters of transcripts sequences of the anti-malarial plant, Artemisia annua". Biotechnology Letters. 34 (4): 737–45. doi:10.1007/s10529-011-0808-0. PMID 22160362. S2CID 14045457.
- ↑ Donaire L, Pedrola L, Rosa R, Llave C (2011). Baxter I (ed.). "High-throughput sequencing of RNA silencing-associated small RNAs in olive (Olea europaea L.)". PLOS ONE. 6 (11): e27916. Bibcode:2011PLoSO...627916D. doi: 10.1371/journal.pone.0027916 . PMC 3225373 . PMID 22140484.
- ↑ Budak H, Akpinar A (November 2011). "Dehydration stress-responsive miRNA in Brachypodium distachyon: evident by genome-wide screening of microRNAs expression". OMICS. 15 (11): 791–9. doi:10.1089/omi.2011.0073. PMID 22122669.
- ↑ Zhang L, Chao JT, Cui MM, Chen YQ, Zong P, Sun YH (July 2011). "[Bioinformatic prediction of conserved microRNAs and their target genes in eggplant (Solanum melongena L.)]". Yi Chuan = Hereditas. 33 (7): 776–84. doi: 10.3724/sp.j.1005.2011.00776 . PMID 22049693.
- ↑ Rajeswaran R, Pooggin MM (January 2012). "RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA". Nucleic Acids Research. 40 (2): 594–9. doi:10.1093/nar/gkr760. PMC 3258149 . PMID 21930511.
- ↑ Wong CE, Zhao YT, Wang XJ, Croft L, Wang ZH, Haerizadeh F, Mattick JS, Singh MB, Carroll BJ, Bhalla PL (May 2011). "MicroRNAs in the shoot apical meristem of soybean". Journal of Experimental Botany. 62 (8): 2495–506. doi:10.1093/jxb/erq437. hdl: 10536/DRO/DU:30106047 . PMID 21504877.
- ↑ Douglas RN, Wiley D, Sarkar A, Springer N, Timmermans MC, Scanlon MJ (May 2010). "ragged seedling2 Encodes an ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves". The Plant Cell. 22 (5): 1441–51. doi:10.1105/tpc.109.071613. PMC 2899878 . PMID 20453116.
- ↑ Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A (April 2010). "miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth". The Plant Cell. 22 (4): 1104–17. doi:10.1105/tpc.109.072553. PMC 2879756 . PMID 20363771.
- ↑ Wang J, Gao X, Li L, Shi X, Zhang J, Shi Z (June 2010). "Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice". Journal of Experimental Botany. 61 (6): 1885–95. doi:10.1093/jxb/erp378. PMC 2852654 . PMID 20080824.
- ↑ Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, Carrington JC (January 2010). "Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing" (PDF). Proceedings of the National Academy of Sciences of the United States of America. 107 (1): 466–71. Bibcode:2010PNAS..107..466C. doi: 10.1073/pnas.0913203107 . PMC 2806713 . PMID 20018656.
- ↑ Yoon EK, Yang JH, Lim J, Kim SH, Kim SK, Lee WS (March 2010). "Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development". Nucleic Acids Research. 38 (4): 1382–91. doi:10.1093/nar/gkp1128. PMC 2831332 . PMID 19969544.
- ↑ Krasnikova MS, Milyutina IA, Bobrova VK, Ozerova LV, Troitsky AV, Solovyev AG, Morozov SY (2009). "Novel miR390-dependent transacting siRNA precursors in plants revealed by a PCR-based experimental approach and database analysis". Journal of Biomedicine & Biotechnology. 2009: 952304. doi: 10.1155/2009/952304 . PMC 2762245 . PMID 19859540.
- ↑ Aly R, Cholakh H, Joel DM, Leibman D, Steinitz B, Zelcer A, Naglis A, Yarden O, Gal-On A (August 2009). "Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant". Plant Biotechnology Journal. 7 (6): 487–98. doi:10.1111/j.1467-7652.2009.00418.x. PMID 19490480.
- ↑ Tedder P, Zubko E, Westhead DR, Meyer P (June 2009). "Small RNA analysis in Petunia hybrida identifies unusual tissue-specific expression patterns of conserved miRNAs and of a 24mer RNA". RNA. 15 (6): 1012–20. doi:10.1261/rna.1517209. PMC 2685514 . PMID 19369427.
- ↑ Li H, Zhang Z, Huang F, Chang L, Ma Y (June 2009). "MicroRNA expression profiles in conventional and micropropagated strawberry (Fragaria x ananassa Duch.) plants". Plant Cell Reports. 28 (6): 891–902. doi:10.1007/s00299-009-0693-3. PMID 19277667. S2CID 10100284.
- ↑ Felippes FF, Weigel D (March 2009). "Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173". EMBO Reports. 10 (3): 264–70. doi:10.1038/embor.2008.247. PMC 2658565 . PMID 19180117.
- ↑ Nogueira FT, Chitwood DH, Madi S, Ohtsu K, Schnable PS, Scanlon MJ, Timmermans MC (January 2009). Copenhaver GP (ed.). "Regulation of small RNA accumulation in the maize shoot apex". PLOS Genetics. 5 (1): e1000320. doi:10.1371/journal.pgen.1000320. PMC 2602737 . PMID 19119413.
- ↑ Abdurakhmonov IY, Devor EJ, Buriev ZT, Huang L, Makamov A, Shermatov SE, Bozorov T, Kushanov FN, Mavlonov GT, Abdukarimov A (September 2008). "Small RNA regulation of ovule development in the cotton plant, G. hirsutum L". BMC Plant Biology. 8: 93. doi:10.1186/1471-2229-8-93. PMC 2564936 . PMID 18793449.
- ↑ Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y (April 2008). "The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins". Plant & Cell Physiology. 49 (4): 493–500. doi: 10.1093/pcp/pcn043 . PMID 18344228.
- ↑ Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC (April 2008). "Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation". Cell. 133 (1): 128–41. doi: 10.1016/j.cell.2008.02.033 . PMID 18342362. S2CID 11174355.
- ↑ Axtell MJ, Snyder JA, Bartel DP (June 2007). "Common functions for diverse small RNAs of land plants". The Plant Cell. 19 (6): 1750–69. doi:10.1105/tpc.107.051706. PMC 1955733 . PMID 17601824.
- ↑ Axtell MJ, Jan C, Rajagopalan R, Bartel DP (November 2006). "A two-hit trigger for siRNA biogenesis in plants". Cell. 127 (3): 565–77. doi: 10.1016/j.cell.2006.09.032 . PMID 17081978. S2CID 1413712.
- ↑ Talmor-Neiman M, Stav R, Klipcan L, Buxdorf K, Baulcombe DC, Arazi T (November 2006). "Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis". The Plant Journal. 48 (4): 511–21. doi: 10.1111/j.1365-313X.2006.02895.x . PMID 17076803.
- ↑ Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC (May 2006). "Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis". Current Biology. 16 (9): 939–44. doi: 10.1016/j.cub.2006.03.065 . PMID 16682356. S2CID 401176.
- ↑ Allen E, Xie Z, Gustafson AM, Carrington JC (April 2005). "microRNA-directed phasing during trans-acting siRNA biogenesis in plants". Cell. 121 (2): 207–21. doi: 10.1016/j.cell.2005.04.004 . PMID 15851028. S2CID 18056486.