The Let-7 microRNA precursor was identified from a study of developmental timing in C. elegans, and was later shown to be part of a much larger class of non-coding RNAs termed microRNAs. miR-98 microRNA precursor from human is a let-7 family member. Let-7 miRNAs have now been predicted or experimentally confirmed in a wide range of species (MIPF0000002). miRNAs are initially transcribed in long transcripts called primary miRNAs (pri-miRNAs), which are processed in the nucleus by Drosha and Pasha to hairpin structures of about 70 nucleotide. These precursors (pre-miRNAs) are exported to the cytoplasm by exportin5, where they are subsequently processed by the enzyme Dicer to a ~22 nucleotide mature miRNA. The involvement of Dicer in miRNA processing demonstrates a relationship with the phenomenon of RNA interference.

The miR-199 microRNA precursor is a short non-coding RNA gene involved in gene regulation. miR-199 genes have now been predicted or experimentally confirmed in mouse, human and a further 21 other species. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The mature products are thought to have regulatory roles through complementarity to mRNA.

There are 89 known sequences today in the microRNA 19 (miR-19) family but it will change quickly. They are found in a large number of vertebrate species. The miR-19 microRNA precursor is a small non-coding RNA molecule that regulates gene expression. Within the human and mouse genome there are three copies of this microRNA that are processed from multiple predicted precursor hairpins:

The miR-1 microRNA precursor is a small micro RNA that regulates its target protein's expression in the cell. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give products at ~22 nucleotides. In this case the mature sequence comes from the 3' arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In humans there are two distinct microRNAs that share an identical mature sequence, and these are called miR-1-1 and miR-1-2.
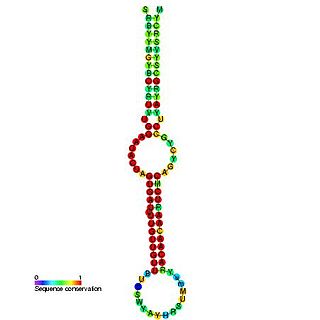
This family represents the microRNA (miRNA) precursor mir-7. This miRNA has been predicted or experimentally confirmed in a wide range of species. miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 5' arm of the precursor. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The involvement of Dicer in miRNA processing suggests a relationship with the phenomenon of RNA interference.

EGF-like domain-containing protein 7 is a protein that in humans is encoded by the EGFL7 gene. Intron 7 of EGFL7 hosts the miR-126 microRNA gene.
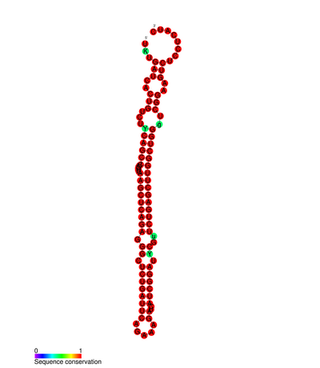
mir-127 microRNA is a short non-coding RNA molecule with interesting overlapping gene structure. miR-127 functions to regulate the expression levels of genes involved in lung development, placental formation and apoptosis. Aberrant expression of miR-127 has been linked to different cancers.

In molecular biology mir-143 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. mir–143 is highly conserved in vertebrates. mir-143 is thought be involved in cardiac morphogenesis but has also been implicated in cancer.

In molecular biology, mir-145 microRNA is a short RNA molecule that in humans is encoded by the MIR145 gene. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
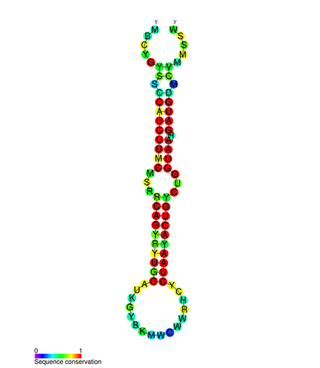
In molecular biology, the miR-200 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by binding and cleaving mRNAs or inhibiting translation. The miR-200 family contains miR-200a, miR-200b, miR-200c, miR-141, and miR-429. There is growing evidence to suggest that miR-200 microRNAs are involved in cancer metastasis.

In molecular biology miR-205 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. They are involved in numerous cellular processes, including development, proliferation, and apoptosis. Currently, it is believed that miRNAs elicit their effect by silencing the expression of target genes.

In molecular biology mir-22 microRNA is a short RNA molecule. MicroRNAs are an abundant class of molecules, approximately 22 nucleotides in length, which can post-transcriptionally regulate gene expression by binding to the 3' UTR of mRNAs expressed in a cell.

In molecular biology mir-451 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

In molecular biology, mir-221 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

In molecular biology mir-210 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

miR-27 is a family of microRNA precursors found in animals, including humans. MicroRNAs are typically transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The excised region or, mature product, of the miR-27 precursor is the microRNA mir-27.

miR-296 is a family of microRNA precursors found in mammals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. This sequence then associates with RISC which effects RNA interference.
In molecular biology mir-301 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-365 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
















