The Let-7 microRNA precursor was identified from a study of developmental timing in C. elegans, and was later shown to be part of a much larger class of non-coding RNAs termed microRNAs. miR-98 microRNA precursor from human is a let-7 family member. Let-7 miRNAs have now been predicted or experimentally confirmed in a wide range of species (MIPF0000002). miRNAs are initially transcribed in long transcripts called primary miRNAs (pri-miRNAs), which are processed in the nucleus by Drosha and Pasha to hairpin structures of about 70 nucleotide. These precursors (pre-miRNAs) are exported to the cytoplasm by exportin5, where they are subsequently processed by the enzyme Dicer to a ~22 nucleotide mature miRNA. The involvement of Dicer in miRNA processing demonstrates a relationship with the phenomenon of RNA interference.
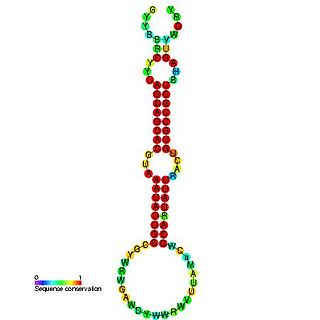
The miR-16 microRNA precursor family is a group of related small non-coding RNA genes that regulates gene expression. miR-16, miR-15, mir-195 and miR-497 are related microRNA precursor sequences from the mir-15 gene family. This microRNA family appears to be vertebrate specific and its members have been predicted or experimentally validated in a wide range of vertebrate species.

The miR-17 microRNA precursor family are a group of related small non-coding RNA genes called microRNAs that regulate gene expression. The microRNA precursor miR-17 family, includes miR-20a/b, miR-93, and miR-106a/b. With the exception of miR-93, these microRNAs are produced from several microRNA gene clusters, which apparently arose from a series of ancient evolutionary genetic duplication events, and also include members of the miR-19, and miR-25 families. These clusters are transcribed as long non-coding RNA transcripts that are processed to form ~70 nucleotide microRNA precursors, that are subsequently processed by the Dicer enzyme to give a ~22 nucleotide products. The mature microRNA products are thought to regulate expression levels of other genes through complementarity to the 3' UTR of specific target messenger RNA.
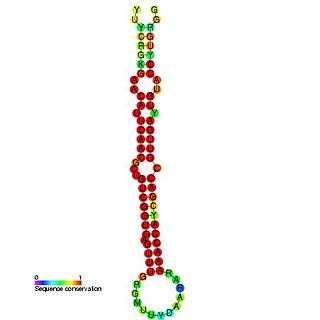
In molecular biology miR-181 microRNA precursor is a small non-coding RNA molecule. MicroRNAs (miRNAs) are transcribed as ~70 nucleotide precursors and subsequently processed by the RNase-III type enzyme Dicer to give a ~22 nucleotide mature product. In this case the mature sequence comes from the 5' arm of the precursor. They target and modulate protein expression by inhibiting translation and / or inducing degradation of target messenger RNAs. This new class of genes has recently been shown to play a central role in malignant transformation. miRNA are downregulated in many tumors and thus appear to function as tumor suppressor genes. The mature products miR-181a, miR-181b, miR-181c or miR-181d are thought to have regulatory roles at posttranscriptional level, through complementarity to target mRNAs. miR-181 which has been predicted or experimentally confirmed in a wide number of vertebrate species as rat, zebrafish, and in the pufferfish.

The miR-29 microRNA precursor, or pre-miRNA, is a small RNA molecule in the shape of a stem-loop or hairpin. Each arm of the hairpin can be processed into one member of a closely related family of short non-coding RNAs that are involved in regulating gene expression. The processed, or "mature" products of the precursor molecule are known as microRNA (miRNA), and have been predicted or confirmed in a wide range of species.
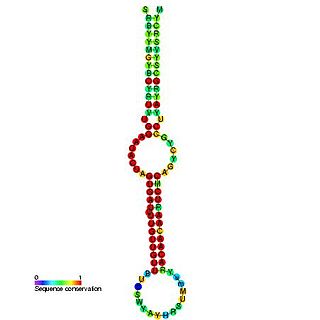
This family represents the microRNA (miRNA) precursor mir-7. This miRNA has been predicted or experimentally confirmed in a wide range of species. miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 5' arm of the precursor. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The involvement of Dicer in miRNA processing suggests a relationship with the phenomenon of RNA interference.

Metastasis-associated protein MTA1 is a protein that in humans is encoded by the MTA1 gene. MTA1 is the founding member of the MTA family of genes. MTA1 is primarily localized in the nucleus but also found to be distributed in the extra-nuclear compartments. MTA1 is a component of several chromatin remodeling complexes including the nucleosome remodeling and deacetylation complex (NuRD). MTA1 regulates gene expression by functioning as a coregulator to integrate DNA-interacting factors to gene activity. MTA1 participates in physiological functions in the normal and cancer cells. MTA1 is one of the most upregulated proteins in human cancer and associates with cancer progression, aggressive phenotypes, and poor prognosis of cancer patients.

Metastasis-associated protein MTA3 is a protein that in humans is encoded by the MTA3 gene. MTA3 protein localizes in the nucleus as well as in other cellular compartments MTA3 is a component of the nucleosome remodeling and deacetylate (NuRD) complex and participates in gene expression. The expression pattern of MTA3 is opposite to that of MTA1 and MTA2 during mammary gland tumorigenesis. However, MTA3 is also overexpressed in a variety of human cancers.

In molecular biology mir-143 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. mir–143 is highly conserved in vertebrates. mir-143 is thought be involved in cardiac morphogenesis but has also been implicated in cancer.
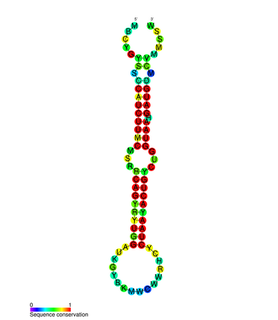
In molecular biology, the miR-200 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by binding and cleaving mRNAs or inhibiting translation. The miR-200 family contains miR-200a, miR-200b, miR-200c, miR-141, and miR-429. There is growing evidence to suggest that miR-200 microRNAs are involved in cancer metastasis.

In molecular biology, mir-433 is a short non-coding RNA molecule. MicroRNAs (miRNAs) function as posttranscriptional regulators of expression levels of other genes by several mechanisms. They play roles in development, metabolism and carcinogenesis.

In molecular biology, mir-221 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
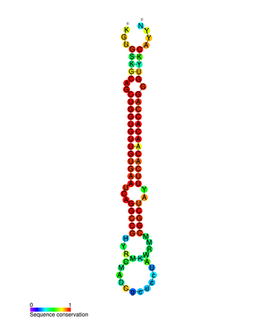
miR-138 is a family of microRNA precursors found in animals, including humans. MicroRNAs are typically transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The excised region or, mature product, of the miR-138 precursor is the microRNA mir-138.
In molecular biology mir-365 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-497 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-153 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-190 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
mir-618 microRNA is a short non-coding RNA molecule belonging both to the family of microRNAs and to that of small interfering RNAs (siRNAs). MicroRNAs function to regulate the expression levels of other genes by several mechanisms, whilst siRNAs are involved primarily with the RNA interference (RNAi) pathway.
In molecular biology mir-765 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.












