Related Research Articles

MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then gene silence said mRNA molecules by one or more of the following processes:
- Cleavage of mRNA strand into two pieces,
- Destabilization of mRNA by shortening its poly(A) tail, or
- Translation of mRNA into proteins.
Small RNA (sRNA) are polymeric RNA molecules that are less than 200 nucleotides in length, and are usually non-coding. RNA silencing is often a function of these molecules, with the most common and well-studied example being RNA interference (RNAi), in which endogenously expressed microRNA (miRNA) or exogenously derived small interfering RNA (siRNA) induces the degradation of complementary messenger RNA. Other classes of small RNA have been identified, including piwi-interacting RNA (piRNA) and its subspecies repeat associated small interfering RNA (rasiRNA). Small RNA "is unable to induce RNAi alone, and to accomplish the task it must form the core of the RNA–protein complex termed the RNA-induced silencing complex (RISC), specifically with Argonaute protein".
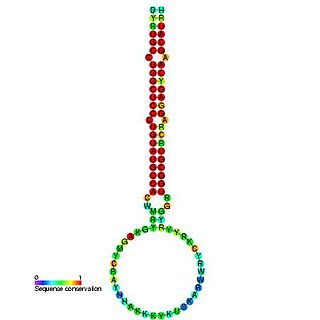
In molecular biology, mir-160 is a microRNA that has been predicted or experimentally confirmed in a range of plant species including Arabidopsis thaliana and Oryza sativa (rice). miR-160 is predicted to bind complementary sites in the untranslated regions of auxin response factor genes to regulate their expression. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.

The mir-172 microRNA is thought to target mRNAs coding for APETALA2-like transcription factors. It has been verified experimentally in the model plant, Arabidopsis thaliana. The mature sequence is excised from the 3' arm of the hairpin.

The miR-17 microRNA precursor family are a group of related small non-coding RNA genes called microRNAs that regulate gene expression. The microRNA precursor miR-17 family, includes miR-20a/b, miR-93, and miR-106a/b. With the exception of miR-93, these microRNAs are produced from several microRNA gene clusters, which apparently arose from a series of ancient evolutionary genetic duplication events, and also include members of the miR-19, and miR-25 families. These clusters are transcribed as long non-coding RNA transcripts that are processed to form ~70 nucleotide microRNA precursors, that are subsequently processed by the Dicer enzyme to give a ~22 nucleotide products. The mature microRNA products are thought to regulate expression levels of other genes through complementarity to the 3' UTR of specific target messenger RNA.
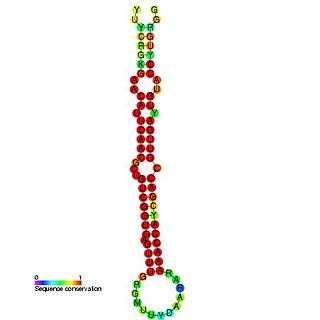
In molecular biology miR-181 microRNA precursor is a small non-coding RNA molecule. MicroRNAs (miRNAs) are transcribed as ~70 nucleotide precursors and subsequently processed by the RNase-III type enzyme Dicer to give a ~22 nucleotide mature product. In this case the mature sequence comes from the 5' arm of the precursor. They target and modulate protein expression by inhibiting translation and / or inducing degradation of target messenger RNAs. This new class of genes has recently been shown to play a central role in malignant transformation. miRNA are downregulated in many tumors and thus appear to function as tumor suppressor genes. The mature products miR-181a, miR-181b, miR-181c or miR-181d are thought to have regulatory roles at posttranscriptional level, through complementarity to target mRNAs. miR-181 which has been predicted or experimentally confirmed in a wide number of vertebrate species as rat, zebrafish, and in the pufferfish.

In molecular biology mir-210 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
Degradome sequencing (Degradome-Seq), also referred to as parallel analysis of RNA ends (PARE), is a modified version of 5'-Rapid Amplification of cDNA Ends (RACE) using high-throughput, deep sequencing methods such as Illumina's SBS technology. The degradome encompasses the entire set of proteases that are expressed at a specific time in a given biological material, including tissues, cells, organisms, and biofluids. Thus, sequencing this degradome offers a method for studying and researching the process of RNA degradation. This process is used to identify and quantify RNA degradation products, or fragments, present in any given biological sample. This approach allows for the systematic identification of targets of RNA decay and provides insight into the dynamics of transcriptional and post-transcriptional gene regulation.

miR-138 is a family of microRNA precursors found in animals, including humans. MicroRNAs are typically transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The excised region or, mature product, of the miR-138 precursor is the microRNA mir-138.
MicroRNA sequencing (miRNA-seq), a type of RNA-Seq, is the use of next-generation sequencing or massively parallel high-throughput DNA sequencing to sequence microRNAs, also called miRNAs. miRNA-seq differs from other forms of RNA-seq in that input material is often enriched for small RNAs. miRNA-seq allows researchers to examine tissue-specific expression patterns, disease associations, and isoforms of miRNAs, and to discover previously uncharacterized miRNAs. Evidence that dysregulated miRNAs play a role in diseases such as cancer has positioned miRNA-seq to potentially become an important tool in the future for diagnostics and prognostics as costs continue to decrease. Like other miRNA profiling technologies, miRNA-Seq has both advantages and disadvantages.
In molecular biology mir-367 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-663 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-390 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-397 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-408 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-474 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-828 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-398 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

DCL1 is a gene in plants that codes for the DCL1 protein, a ribonuclease III enzyme involved in processing double-stranded RNA (dsRNA) and microRNA (miRNA). Although DCL1, also called Endoribonuclease Dicer homolog 1, is named for its homology with the metazoan protein Dicer, its role in miRNA biogenesis is somewhat different, due to substantial differences in miRNA maturation processes between plants and animals, as well due to additional downstream plant-specific pathways, where DCL1 paralogs like DCL4 participate, such Trans-acting siRNA biogenesis.
Jian-Kang Zhu is a plant scientist, researcher and academic. He is a Senior Principal Investigator in the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences (CAS). He is also the Academic Director of CAS Center of Excellence in Plant Sciences.