
Catenins are a family of proteins found in complexes with cadherin cell adhesion molecules of animal cells. The first two catenins that were identified became known as α-catenin and β-catenin. α-Catenin can bind to β-catenin and can also bind filamentous actin (F-actin). β-Catenin binds directly to the cytoplasmic tail of classical cadherins. Additional catenins such as γ-catenin and δ-catenin have been identified. The name "catenin" was originally selected because it was suspected that catenins might link cadherins to the cytoskeleton.
The epithelial–mesenchymal transition (EMT) is a process by which epithelial cells lose their cell polarity and cell–cell adhesion, and gain migratory and invasive properties to become mesenchymal stem cells; these are multipotent stromal cells that can differentiate into a variety of cell types. EMT is essential for numerous developmental processes including mesoderm formation and neural tube formation. EMT has also been shown to occur in wound healing, in organ fibrosis and in the initiation of metastasis in cancer progression.

Mesenchyme is a type of loosely organized animal embryonic connective tissue of undifferentiated cells that give rise to most tissues, such as skin, blood or bone. The interactions between mesenchyme and epithelium help to form nearly every organ in the developing embryo.

The miR-9 microRNA, is a short non-coding RNA gene involved in gene regulation. The mature ~21nt miRNAs are processed from hairpin precursor sequences by the Dicer enzyme. The dominant mature miRNA sequence is processed from the 5' arm of the mir-9 precursor, and from the 3' arm of the mir-79 precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In vertebrates, miR-9 is highly expressed in the brain, and is suggested to regulate neuronal differentiation. A number of specific targets of miR-9 have been proposed, including the transcription factor REST and its partner CoREST.

Kruppel-like factor 4 is a member of the KLF family of zinc finger transcription factors, which belongs to the relatively large family of SP1-like transcription factors. KLF4 is involved in the regulation of proliferation, differentiation, apoptosis and somatic cell reprogramming. Evidence also suggests that KLF4 is a tumor suppressor in certain cancers, including colorectal cancer. It has three C2H2-zinc fingers at its carboxyl terminus that are closely related to another KLF, KLF2. It has two nuclear localization sequences that signals it to localize to the nucleus. In embryonic stem cells (ESCs), KLF4 has been demonstrated to be a good indicator of stem-like capacity. It is suggested that the same is true in mesenchymal stem cells (MSCs).
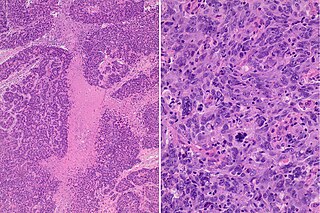
The basal-like carcinoma is a recently proposed subtype of breast cancer defined by its gene expression and protein expression profile.

Epithelial cell adhesion molecule (EpCAM), also known as CD326 among other names, is a transmembrane glycoprotein mediating Ca2+-independent homotypic cell–cell adhesion in epithelia. EpCAM is also involved in cell signaling, migration, proliferation, and differentiation. Additionally, EpCAM has oncogenic potential via its capacity to upregulate c-myc, e-fabp, and cyclins A & E. Since EpCAM is expressed exclusively in epithelia and epithelial-derived neoplasms, EpCAM can be used as diagnostic marker for various cancers. It appears to play a role in tumorigenesis and metastasis of carcinomas, so it can also act as a potential prognostic marker and as a potential target for immunotherapeutic strategies.

Zinc finger protein SNAI1 is a protein that in humans is encoded by the SNAI1 gene. Snail is a family of transcription factors that promote the repression of the adhesion molecule E-cadherin to regulate epithelial to mesenchymal transition (EMT) during embryonic development.
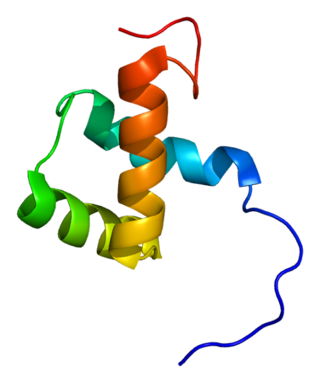
Zinc finger E-box-binding homeobox 1 is a protein that in humans is encoded by the ZEB1 gene.

Metastasis-associated protein MTA2 is a protein that in humans is encoded by the MTA2 gene.

Metastasis-associated protein MTA3 is a protein that in humans is encoded by the MTA3 gene. MTA3 protein localizes in the nucleus as well as in other cellular compartments MTA3 is a component of the nucleosome remodeling and deacetylate (NuRD) complex and participates in gene expression. The expression pattern of MTA3 is opposite to that of MTA1 and MTA2 during mammary gland tumorigenesis. However, MTA3 is also overexpressed in a variety of human cancers.

Cadherin-1 or Epithelial cadherin(E-cadherin), is a protein that in humans is encoded by the CDH1 gene. Mutations are correlated with gastric, breast, colorectal, thyroid, and ovarian cancers. CDH1 has also been designated as CD324. It is a tumor suppressor gene.
A mesenchymal–epithelial transition (MET) is a reversible biological process that involves the transition from motile, multipolar or spindle-shaped mesenchymal cells to planar arrays of polarized cells called epithelia. MET is the reverse process of epithelial–mesenchymal transition (EMT) and it has been shown to occur in normal development, induced pluripotent stem cell reprogramming, cancer metastasis and wound healing.

In molecular biology miR-203 is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms, such as translational repression and Argonaute-catalyzed messenger RNA cleavage. miR-203 has been identified as a skin-specific microRNA, and it forms an expression gradient that defines the boundary between proliferative epidermal basal progenitors and terminally differentiating suprabasal cells. It has also been found upregulated in psoriasis and differentially expressed in some types of cancer.

In molecular biology miR-205 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. They are involved in numerous cellular processes, including development, proliferation, and apoptosis. Currently, it is believed that miRNAs elicit their effect by silencing the expression of target genes.

miR-138 is a family of microRNA precursors found in animals, including humans. MicroRNAs are typically transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The excised region or, mature product, of the miR-138 precursor is the microRNA mir-138.
In molecular biology mir-661 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
Migration inducting gene 7 is a gene that corresponds to a cysteine-rich protein localized to the cell membrane and cytoplasm. It is the first-in-class of novel proteins translated from what are thought to be long Non-coding RNAs.

MicroRNA 200c is a microRNA that in humans is encoded by the MIR200C gene.
For cancer, invasion is the direct extension and penetration by cancer cells into neighboring tissues. It is generally distinguished from metastasis, which is the spread of cancer cells through the circulatory system or the lymphatic system to more distant locations. Yet, lymphovascular invasion is generally the first step of metastasis.















