Malignant transformation is the process by which cells acquire the properties of cancer. This may occur as a primary process in normal tissue, or secondarily as malignant degeneration of a previously existing benign tumor.
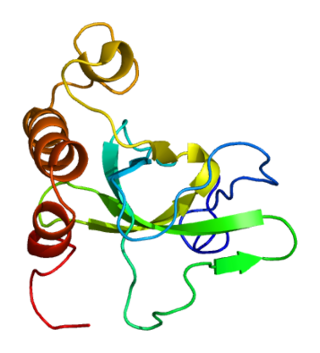
MSH6 or mutS homolog 6 is a gene that codes for DNA mismatch repair protein Msh6 in the budding yeast Saccharomyces cerevisiae. It is the homologue of the human "G/T binding protein," (GTBP) also called p160 or hMSH6. The MSH6 protein is a member of the Mutator S (MutS) family of proteins that are involved in DNA damage repair.

The miR-17 microRNA precursor family are a group of related small non-coding RNA genes called microRNAs that regulate gene expression. The microRNA precursor miR-17 family, includes miR-20a/b, miR-93, and miR-106a/b. With the exception of miR-93, these microRNAs are produced from several microRNA gene clusters, which apparently arose from a series of ancient evolutionary genetic duplication events, and also include members of the miR-19, and miR-25 families. These clusters are transcribed as long non-coding RNA transcripts that are processed to form ~70 nucleotide microRNA precursors, that are subsequently processed by the Dicer enzyme to give a ~22 nucleotide products. The mature microRNA products are thought to regulate expression levels of other genes through complementarity to the 3' UTR of specific target messenger RNA.

There are 89 known sequences today in the microRNA 19 (miR-19) family but it will change quickly. They are found in a large number of vertebrate species. The miR-19 microRNA precursor is a small non-coding RNA molecule that regulates gene expression. Within the human and mouse genome there are three copies of this microRNA that are processed from multiple predicted precursor hairpins:

The miR-1 microRNA precursor is a small micro RNA that regulates its target protein's expression in the cell. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give products at ~22 nucleotides. In this case the mature sequence comes from the 3' arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In humans there are two distinct microRNAs that share an identical mature sequence, and these are called miR-1-1 and miR-1-2.

microRNA 21 also known as hsa-mir-21 or miRNA21 is a mammalian microRNA that is encoded by the MIR21 gene.
An oncomir is a microRNA (miRNA) that is associated with cancer. MicroRNAs are short RNA molecules about 22 nucleotides in length. Essentially, miRNAs specifically target certain messenger RNAs (mRNAs) to prevent them from coding for a specific protein. The dysregulation of certain microRNAs (oncomirs) has been associated with specific cancer forming (oncogenic) events. Many different oncomirs have been identified in numerous types of human cancers.

In molecular biology mir-126 is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several pre- and post-transcription mechanisms.
mir-127 microRNA is a short non-coding RNA molecule with interesting overlapping gene structure. miR-127 functions to regulate the expression levels of genes involved in lung development, placental formation and apoptosis. Aberrant expression of miR-127 has been linked to different cancers.

In molecular biology, miR-137 is a short non-coding RNA molecule that functions to regulate the expression levels of other genes by various mechanisms. miR-137 is located on human chromosome 1p22 and has been implicated to act as a tumor suppressor in several cancer types including colorectal cancer, squamous cell carcinoma and melanoma via cell cycle control.

In molecular biology, miR-184 microRNA is a short non-coding RNA molecule. MicroRNAs (miRNAs) function as posttranscriptional regulators of expression levels of other genes by several mechanisms. Several targets for miR-184 have been described, including that of mediators of neurological development, apoptosis and it has been suggested that miR-184 plays an essential role in development.
In molecular biology, the miR-200 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by binding and cleaving mRNAs or inhibiting translation. The miR-200 family contains miR-200a, miR-200b, miR-200c, miR-141, and miR-429. There is growing evidence to suggest that miR-200 microRNAs are involved in cancer metastasis.

In molecular biology miR-203 is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms, such as translational repression and Argonaute-catalyzed messenger RNA cleavage. miR-203 has been identified as a skin-specific microRNA, and it forms an expression gradient that defines the boundary between proliferative epidermal basal progenitors and terminally differentiating suprabasal cells. It has also been found upregulated in psoriasis and differentially expressed in some types of cancer.

In molecular biology miR-205 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. They are involved in numerous cellular processes, including development, proliferation, and apoptosis. Currently, it is believed that miRNAs elicit their effect by silencing the expression of target genes.
miR-31 has been characterised as a tumour suppressor miRNA, with its levels varying in breast cancer cells according to the metastatic state of the tumour. From its typical abundance in healthy tissue is a moderate decrease in non-metastatic breast cancer cell lines, and levels are almost completely absent in mouse and human metastatic breast cancer cell lines. Mir-31-5p has also been observed upregulated in Zinc Deficient rats compared to normal in ESCC and in other types of cancers when using this animal model. There has also been observed a strong encapsulation of tumour cells expressing miR-31, as well as a reduced cell survival rate. miR-31's antimetastatic effects therefore make it a potential therapeutic target for breast cancer. However, these two papers were formally retracted by the authors in 2015.

HOXA11-AS lncRNA is a long non-coding RNA from the antisense strand in the homeobox A. The HOX gene contains four clusters. The sense strand of the HOXA gene codes for proteins. Alternative names for HOXA11-AS lncRNA are: HOXA-AS5, HOXA11S, HOXA11-AS1, HOXA11AS, or NCRNA00076. This gene is 3,885 nucleotides long and resides at chromosome 7 (7p15.2) and is transcribed from an independent gene promoter. Being a lncRNA, it is longer than 200 nucleotides in length, in contrast to regular non-coding RNAs.

Cancer epigenetics is the study of epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of expression of genes that occurs about 10 times more frequently by transcription silencing than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations. However, in colon tumors compared to adjacent normal-appearing colonic mucosa, there are about 600 to 800 heavily methylated CpG islands in the promoters of genes in the tumors while these CpG islands are not methylated in the adjacent mucosa. Manipulation of epigenetic alterations holds great promise for cancer prevention, detection, and therapy. In different types of cancer, a variety of epigenetic mechanisms can be perturbed, such as the silencing of tumor suppressor genes and activation of oncogenes by altered CpG island methylation patterns, histone modifications, and dysregulation of DNA binding proteins. There are several medications which have epigenetic impact, that are now used in a number of these diseases.
In molecular biology mir-186 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
DNA methylation in cancer plays a variety of roles, helping to change the healthy cells by regulation of gene expression to a cancer cells or a diseased cells disease pattern. One of the most widely studied DNA methylation dysregulation is the promoter hypermethylation where the CPGs islands in the promoter regions are methylated contributing or causing genes to be silenced.















