
MicroRNA (miRNA) precursor miR156 is a family of plant non-coding RNA. This microRNA has now been predicted or experimentally confirmed in a range of plant species. Animal miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. miR156 functions in the induction of flowering by suppressing the transcripts of SQUAMOSA-PROMOTER BINDING LIKE (SPL) transcription factors gene family. It was suggested that the loading into ARGONAUTE1 and ARGONAUTE5 is required for miR156 functionality in Arabidopsis thaliana. In plants the precursor sequences may be longer, and the carpel factory (caf) enzyme appears to be involved in processing. In this case the mature sequence comes from the 5' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complementarity to mRNA.
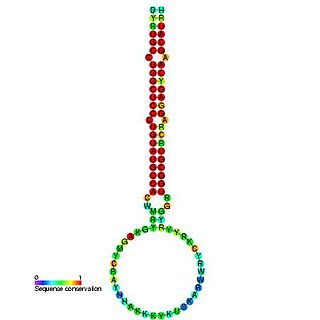
In molecular biology, mir-160 is a microRNA that has been predicted or experimentally confirmed in a range of plant species including Arabidopsis thaliana and Oryza sativa (rice). miR-160 is predicted to bind complementary sites in the untranslated regions of auxin response factor genes to regulate their expression. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.
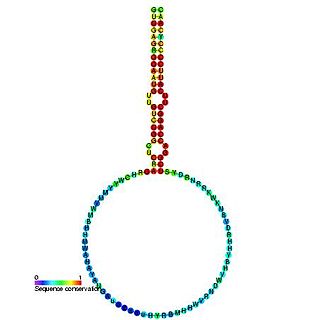
The plant mir-166 microRNA precursor is a small non-coding RNA gene. This microRNA (miRNA) has now been predicted or experimentally confirmed in a wide range of plant species. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 3' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The mature products are thought to have regulatory roles through complementarity to messenger RNA.

mir-395 is a non-coding RNA called a microRNA that was identified in both Arabidopsis thaliana and Oryza sativa computationally and was later experimentally verified. mir-395 is thought to target mRNAs coding for ATP sulphurylases. The mature sequence is excised from the 3' arm of the hairpin.

In molecular biology, Small nucleolar RNA Z151 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z151 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs Plant snoRNA Z151 was identified in screens of Oryza sativa and Arabidopsis thaliana.

mir-399 is a microRNA that was identified in both Arabidopsis thaliana and Oryza sativa computationally and was later experimentally verified. mir-399 is thought to target mRNAs coding for a phosphate transporter. The mature sequence is excised from the 3' arm of the hairpin. There are multiple copies of MIR399 in each plant genome, for example A. thaliana contains six microRNA precursors that all give rise to an almost identical mature miR-399 sequence.
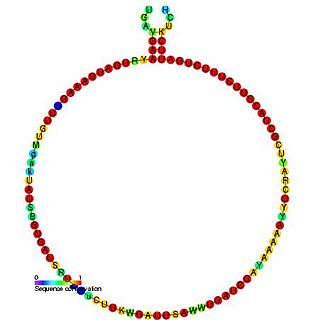
In molecular biology, Small nucleolar RNA Z41 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z41 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA Z41 was identified in screens of Arabidopsis thaliana.
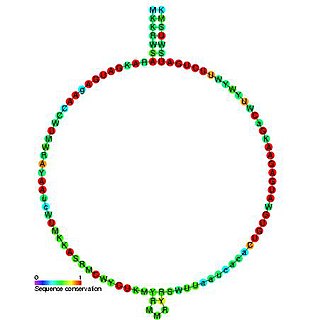
In molecular biology, Small nucleolar RNA Z110 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z110 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs Plant snoRNA Z110 was identified in screens of Arabidopsis thaliana and Oryza sativa .

In molecular biology, Small nucleolar RNA RZ107/R87 refers to a group of related non-coding RNA (ncRNA) molecules which function in the biogenesis of other small nuclear RNAs (snRNAs). These small nucleolar RNAs (snoRNAs) are modifying RNAs and usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis.
In molecular biology, Small nucleolar RNA R20 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA R20 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA R20 was identified in a screen of Arabidopsis thaliana.
In molecular biology, the Small nucleolar RNA snoR1 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snoR1 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA snoR1 was identified in a screen of Arabidopsis thaliana.

In molecular biology, Small nucleolar RNA R28 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA R28 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA R28 was identified in a screen of Arabidopsis thaliana.

In molecular biology, Small nucleolar RNA Z157 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z157 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs.

In molecular biology, SNORD15 is a non-coding RNA (ncRNA) molecule which functions in the modification of small nuclear RNAs. This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, Small nucleolar RNA Z152 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z152 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA Z152 was identified in screens of Oryza sativa and Arabidopsis thaliana.
Trans-acting siRNA are a class of small interfering RNA (siRNA) that repress gene expression through post-transcriptional gene silencing in land plants. Precursor transcripts from TAS loci are polyadenylated and converted to double-stranded RNA, and are then processed into 21-nucleotide-long RNA duplexes with overhangs. These segments are incorporated into an RNA-induced silencing complex (RISC) and direct the sequence-specific cleavage of target mRNA. Ta-siRNAs are classified as siRNA because they arise from double-stranded RNA (dsRNA).
In molecular biology mir-390 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-396 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-398 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
Jian-Kang Zhu is a plant scientist, researcher and academic. He is a Senior Principal Investigator in the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences (CAS). He is also the Academic Director of CAS Center of Excellence in Plant Sciences.















