The Let-7 microRNA precursor was identified from a study of developmental timing in C. elegans, and was later shown to be part of a much larger class of non-coding RNAs termed microRNAs. miR-98 microRNA precursor from human is a let-7 family member. Let-7 miRNAs have now been predicted or experimentally confirmed in a wide range of species (MIPF0000002). miRNAs are initially transcribed in long transcripts called primary miRNAs (pri-miRNAs), which are processed in the nucleus by Drosha and Pasha to hairpin structures of about 70 nucleotide. These precursors (pre-miRNAs) are exported to the cytoplasm by exportin5, where they are subsequently processed by the enzyme Dicer to a ~22 nucleotide mature miRNA. The involvement of Dicer in miRNA processing demonstrates a relationship with the phenomenon of RNA interference.

The miR-103 microRNA precursor, is a short non-coding RNA gene involved in gene regulation. miR-103 and miR-107 have now been predicted or experimentally confirmed in human.
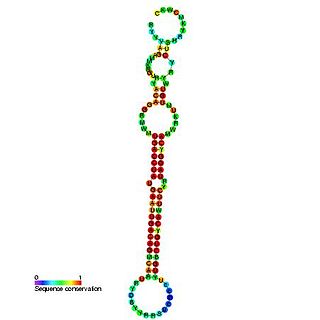
The miR-192 microRNA precursor, is a short non-coding RNA gene involved in gene regulation. miR-192 and miR-215 have now been predicted or experimentally confirmed in mouse and human.

MicroRNA (miRNA) precursor miR156 is a family of plant non-coding RNA. This microRNA has now been predicted or experimentally confirmed in a range of plant species. Animal miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. miR156 functions in the induction of flowering by suppressing the transcripts of SQUAMOSA-PROMOTER BINDING LIKE (SPL) transcription factors gene family. It was suggested that the loading into ARGONAUTE1 and ARGONAUTE5 is required for miR156 functionality in Arabidopsis thaliana. In plants the precursor sequences may be longer, and the carpel factory (caf) enzyme appears to be involved in processing. In this case the mature sequence comes from the 5' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complementarity to mRNA.
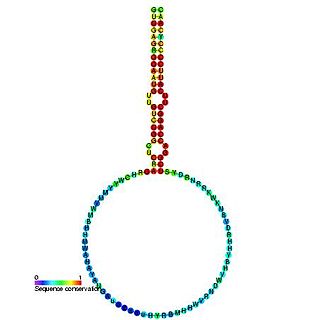
The plant mir-166 microRNA precursor is a small non-coding RNA gene. This microRNA (miRNA) has now been predicted or experimentally confirmed in a wide range of plant species. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 3' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The mature products are thought to have regulatory roles through complementarity to messenger RNA.

The miR-199 microRNA precursor is a short non-coding RNA gene involved in gene regulation. miR-199 genes have now been predicted or experimentally confirmed in mouse, human and a further 21 other species. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The mature products are thought to have regulatory roles through complementarity to mRNA.

The miR-29 microRNA precursor, or pre-miRNA, is a small RNA molecule in the shape of a stem-loop or hairpin. Each arm of the hairpin can be processed into one member of a closely related family of short non-coding RNAs that are involved in regulating gene expression. The processed, or "mature" products of the precursor molecule are known as microRNA (miRNA), and have been predicted or confirmed in a wide range of species.
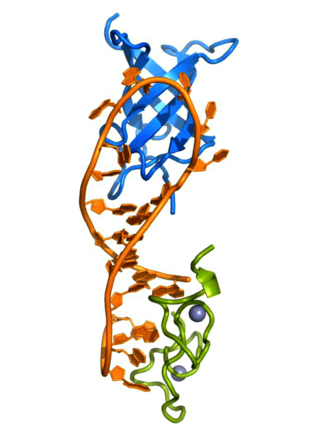
Lin-28 homolog A is a protein that in humans is encoded by the LIN28 gene.

Victor R. Ambros is an American developmental biologist who discovered the first known microRNA (miRNA). He is a professor at the University of Massachusetts Medical School in Worcester, Massachusetts.
Gary Bruce Ruvkun is an American molecular biologist at Massachusetts General Hospital and professor of genetics at Harvard Medical School in Boston. Ruvkun discovered the mechanism by which lin-4, the first microRNA (miRNA) discovered by Victor Ambros, regulates the translation of target messenger RNAs via imperfect base-pairing to those targets, and discovered the second miRNA, let-7, and that it is conserved across animal phylogeny, including in humans. These miRNA discoveries revealed a new world of RNA regulation at an unprecedented small size scale, and the mechanism of that regulation. Ruvkun also discovered many features of insulin-like signaling in the regulation of aging and metabolism. He was elected a Member of the American Philosophical Society in 2019.

miR-224 is a family of microRNA precursors found in mammals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer.

miR-144 is a family of microRNA precursors found in mammals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. In humans, miR-144 has been characterised as a "common miRNA signature" of a number of different tumours.

miR-338 is a family of brain-specific microRNA precursors found in mammals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. This sequence then associates with RISC which effects RNA interference.

miR-150 is a family of microRNA precursors found in mammals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. This sequence then associates with RISC which effects RNA interference.

miR-191 is a family of microRNA precursors found in mammals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. This sequence then associates with RISC which effects RNA interference.

miR-208 is a family of microRNA precursors found in animals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. This sequence then associates with RISC which effects RNA interference.

miR-296 is a family of microRNA precursors found in mammals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. This sequence then associates with RISC which effects RNA interference.
mir-48 microRNA is a microRNA which is found in nematodes, in which it controls developmental timing. It acts in the heterochronic pathway, where it controls the timing of cell fate decisions in the vulva and hypodermis during larval development.
In molecular biology mir-84 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-241 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.















