
MicroRNA (miRNA) are small, single-stranded, non-coding RNA molecules containing 21 to 23 nucleotides. Found in plants, animals and some viruses, miRNAs are involved in RNA silencing and post-transcriptional regulation of gene expression. miRNAs base-pair to complementary sequences in mRNA molecules, then gene silence said mRNA molecules by one or more of the following processes:
- Cleavage of mRNA strand into two pieces,
- Destabilization of mRNA by shortening its poly(A) tail, or
- Translation of mRNA into proteins.
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either transcription or translation and is often used in research. In particular, methods used to silence genes are being increasingly used to produce therapeutics to combat cancer and other diseases, such as infectious diseases and neurodegenerative disorders.

Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.

The miR-9 microRNA, is a short non-coding RNA gene involved in gene regulation. The mature ~21nt miRNAs are processed from hairpin precursor sequences by the Dicer enzyme. The dominant mature miRNA sequence is processed from the 5' arm of the mir-9 precursor, and from the 3' arm of the mir-79 precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In vertebrates, miR-9 is highly expressed in the brain, and is suggested to regulate neuronal differentiation. A number of specific targets of miR-9 have been proposed, including the transcription factor REST and its partner CoREST.

mir-133 is a type of non-coding RNA called a microRNA that was first experimentally characterised in mice. Homologues have since been discovered in several other species including invertebrates such as the fruitfly Drosophila melanogaster. Each species often encodes multiple microRNAs with identical or similar mature sequence. For example, in the human genome there are three known miR-133 genes: miR-133a-1, miR-133a-2 and miR-133b found on chromosomes 18, 20 and 6 respectively. The mature sequence is excised from the 3' arm of the hairpin. miR-133 is expressed in muscle tissue and appears to repress the expression of non-muscle genes.
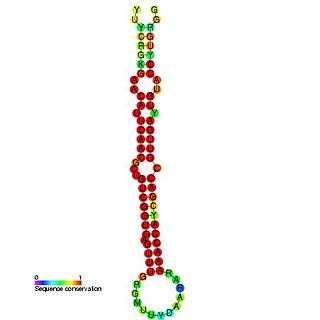
In molecular biology miR-181 microRNA precursor is a small non-coding RNA molecule. MicroRNAs (miRNAs) are transcribed as ~70 nucleotide precursors and subsequently processed by the RNase-III type enzyme Dicer to give a ~22 nucleotide mature product. In this case the mature sequence comes from the 5' arm of the precursor. They target and modulate protein expression by inhibiting translation and / or inducing degradation of target messenger RNAs. This new class of genes has recently been shown to play a central role in malignant transformation. miRNA are downregulated in many tumors and thus appear to function as tumor suppressor genes. The mature products miR-181a, miR-181b, miR-181c or miR-181d are thought to have regulatory roles at posttranscriptional level, through complementarity to target mRNAs. miR-181 which has been predicted or experimentally confirmed in a wide number of vertebrate species as rat, zebrafish, and in the pufferfish.

There are 89 known sequences today in the microRNA 19 (miR-19) family but it will change quickly. They are found in a large number of vertebrate species. The miR-19 microRNA precursor is a small non-coding RNA molecule that regulates gene expression. Within the human and mouse genome there are three copies of this microRNA that are processed from multiple predicted precursor hairpins:

The miR-1 microRNA precursor is a small micro RNA that regulates its target protein's expression in the cell. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give products at ~22 nucleotides. In this case the mature sequence comes from the 3' arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In humans there are two distinct microRNAs that share an identical mature sequence, and these are called miR-1-1 and miR-1-2.
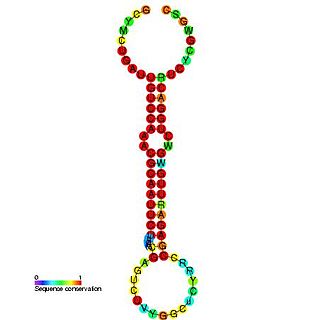
In molecular biology, the microRNA miR-219 was predicted in vertebrates by conservation between human, mouse and pufferfish and cloned in pufferfish. It was later predicted and confirmed experimentally in Drosophila. Homologs of miR-219 have since been predicted or experimentally confirmed in a wide range of species, including the platyhelminth Schmidtea mediterranea, several arthropod species and a wide range of vertebrates. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.
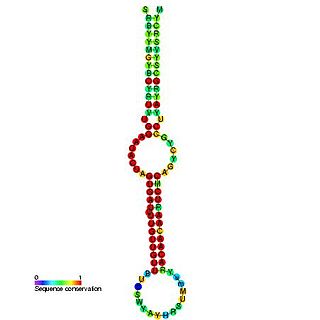
This family represents the microRNA (miRNA) precursor mir-7. This miRNA has been predicted or experimentally confirmed in a wide range of species. miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 5' arm of the precursor. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The involvement of Dicer in miRNA processing suggests a relationship with the phenomenon of RNA interference.

microRNA 21 also known as hsa-mir-21 or miRNA21 is a mammalian microRNA that is encoded by the MIR21 gene.

miR-96 microRNA precursor is a small non-coding RNA that regulates gene expression. microRNAs are transcribed as ~80 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~23 nucleotide products. In this case the mature sequence comes from the 5′ arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA.

MiR-155 is a microRNA that in humans is encoded by the MIR155 host gene or MIR155HG. MiR-155 plays a role in various physiological and pathological processes. Exogenous molecular control in vivo of miR-155 expression may inhibit malignant growth, viral infections, and enhance the progression of cardiovascular diseases.

In molecular biology mir-126 is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several pre- and post-transcription mechanisms.
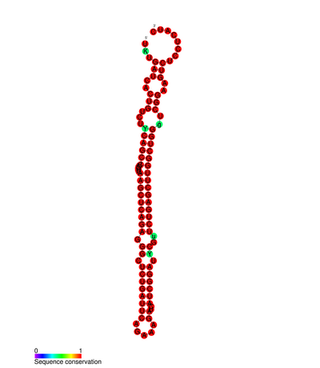
mir-127 microRNA is a short non-coding RNA molecule with interesting overlapping gene structure. miR-127 functions to regulate the expression levels of genes involved in lung development, placental formation and apoptosis. Aberrant expression of miR-127 has been linked to different cancers.

In molecular biology mir-143 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. mir–143 is highly conserved in vertebrates. mir-143 is thought be involved in cardiac morphogenesis but has also been implicated in cancer.

In molecular biology mir-22 microRNA is a short RNA molecule. MicroRNAs are an abundant class of molecules, approximately 22 nucleotides in length, which can post-transcriptionally regulate gene expression by binding to the 3' UTR of mRNAs expressed in a cell.

miR-296 is a family of microRNA precursors found in mammals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. This sequence then associates with RISC which effects RNA interference.
lsy-6 microRNA belongs to the class of miRNAs; these function to regulate the expression levels of other genes by several mechanisms. lsy-6 is a short non-coding RNA molecule and the first miRNA identified as having a role in nervous system development. It regulates left-right neuronal asymmetry in the nematode worm Caenorhabditis elegans.
In molecular biology mir-153 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

















