
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products. Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network.
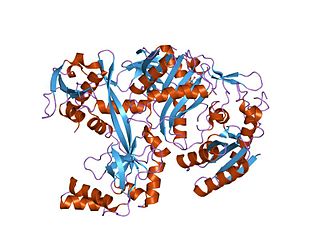
Piwi genes were identified as regulatory proteins responsible for stem cell and germ cell differentiation. Piwi is an abbreviation of P-elementInduced WImpy testis in Drosophila. Piwi proteins are highly conserved RNA-binding proteins and are present in both plants and animals. Piwi proteins belong to the Argonaute/Piwi family and have been classified as nuclear proteins. Studies on Drosophila have also indicated that Piwi proteins have no slicer activity conferred by the presence of the Piwi domain. In addition, Piwi associates with heterochromatin protein 1, an epigenetic modifier, and piRNA-complementary sequences. These are indications of the role Piwi plays in epigenetic regulation. Piwi proteins are also thought to control the biogenesis of piRNA as many Piwi-like proteins contain slicer activity which would allow Piwi proteins to process precursor piRNA into mature piRNA.
The Let-7 microRNA precursor was identified from a study of developmental timing in C. elegans, and was later shown to be part of a much larger class of non-coding RNAs termed microRNAs. miR-98 microRNA precursor from human is a let-7 family member. Let-7 miRNAs have now been predicted or experimentally confirmed in a wide range of species (MIPF0000002). miRNAs are initially transcribed in long transcripts called primary miRNAs (pri-miRNAs), which are processed in the nucleus by Drosha and Pasha to hairpin structures of about 70 nucleotide. These precursors (pre-miRNAs) are exported to the cytoplasm by exportin5, where they are subsequently processed by the enzyme Dicer to a ~22 nucleotide mature miRNA. The involvement of Dicer in miRNA processing demonstrates a relationship with the phenomenon of RNA interference.

The miR-9 microRNA, is a short non-coding RNA gene involved in gene regulation. The mature ~21nt miRNAs are processed from hairpin precursor sequences by the Dicer enzyme. The dominant mature miRNA sequence is processed from the 5' arm of the mir-9 precursor, and from the 3' arm of the mir-79 precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In vertebrates, miR-9 is highly expressed in the brain, and is suggested to regulate neuronal differentiation. A number of specific targets of miR-9 have been proposed, including the transcription factor REST and its partner CoREST.

The mir-10 microRNA precursor is a short non-coding RNA gene involved in gene regulation. It is part of an RNA gene family which contains mir-10, mir-51, mir-57, mir-99 and mir-100. mir-10, mir-99 and mir-100 have now been predicted or experimentally confirmed in a wide range of species. miR-51 and miR-57 have currently only been identified in the nematode Caenorhabditis elegans.

The miR-129 microRNA precursor is a small non-coding RNA molecule that regulates gene expression. This microRNA was first experimentally characterised in mouse and homologues have since been discovered in several other species, such as humans, rats and zebrafish. The mature sequence is excised by the Dicer enzyme from the 5' arm of the hairpin. It was elucidated by Calin et al. that miR-129-1 is located in a fragile site region of the human genome near a specific site, FRA7H in chromosome 7q32, which is a site commonly deleted in many cancers. miR-129-2 is located in 11p11.2.

In molecular biology, miR-130 microRNA precursor is a small non-coding RNA that regulates gene expression. This microRNA has been identified in mouse, and in human. miR-130 appears to be vertebrate-specific miRNA and has now been predicted or experimentally confirmed in a range of vertebrate species. Mature microRNAs are processed from the precursor stem-loop by the Dicer enzyme. In this case, the mature sequence is excised from the 3' arm of the hairpin. It has been found that miR-130 is upregulated in a type of cancer called hepatocellular carcinoma. It has been shown that miR-130a is expressed in the hematopoietic stem/progenitor cell compartment but not in mature blood cells.

There are 89 known sequences today in the microRNA 19 (miR-19) family but it will change quickly. They are found in a large number of vertebrate species. The miR-19 microRNA precursor is a small non-coding RNA molecule that regulates gene expression. Within the human and mouse genome there are three copies of this microRNA that are processed from multiple predicted precursor hairpins:

The miR-1 microRNA precursor is a small micro RNA that regulates its target protein's expression in the cell. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give products at ~22 nucleotides. In this case the mature sequence comes from the 3' arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In humans there are two distinct microRNAs that share an identical mature sequence, and these are called miR-1-1 and miR-1-2.
The miR-34 microRNA precursor family are non-coding RNA molecules that, in mammals, give rise to three major mature miRNAs. The miR-34 family members were discovered computationally and later verified experimentally. The precursor miRNA stem-loop is processed in the cytoplasm of the cell, with the predominant miR-34 mature sequence excised from the 5' arm of the hairpin.

Methyl-CpG-binding domain protein 3 is a protein that in humans is encoded by the MBD3 gene.

In molecular biology, miR-137 is a short non-coding RNA molecule that functions to regulate the expression levels of other genes by various mechanisms. miR-137 is located on human chromosome 1p22 and has been implicated to act as a tumor suppressor in several cancer types including colorectal cancer, squamous cell carcinoma and melanoma via cell cycle control.
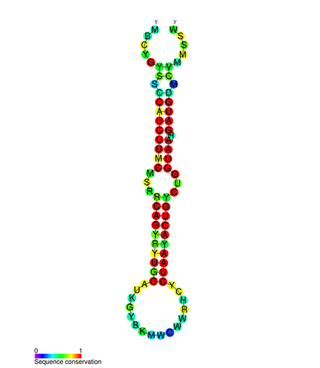
In molecular biology, the miR-200 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by binding and cleaving mRNAs or inhibiting translation. The miR-200 family contains miR-200a, miR-200b, miR-200c, miR-141, and miR-429. There is growing evidence to suggest that miR-200 microRNAs are involved in cancer metastasis.

In molecular biology miR-203 is a short non-coding RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms, such as translational repression and Argonaute-catalyzed messenger RNA cleavage. miR-203 has been identified as a skin-specific microRNA, and it forms an expression gradient that defines the boundary between proliferative epidermal basal progenitors and terminally differentiating suprabasal cells. It has also been found upregulated in psoriasis and differentially expressed in some types of cancer.

In molecular biology miR-205 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. They are involved in numerous cellular processes, including development, proliferation, and apoptosis. Currently, it is believed that miRNAs elicit their effect by silencing the expression of target genes.
High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) is a variant of CLIP for genome-wide mapping protein–RNA binding sites or RNA modification sites in vivo. HITS-CLIP was originally used to generate genome-wide protein-RNA interaction maps for the neuron-specific RNA-binding protein and splicing factor NOVA1 and NOVA2; since then a number of other splicing factor maps have been generated, including those for PTB, RbFox2, SFRS1, hnRNP C, and even N6-Methyladenosine (m6A) mRNA modifications.

miR-138 is a family of microRNA precursors found in animals, including humans. MicroRNAs are typically transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The excised region or, mature product, of the miR-138 precursor is the microRNA mir-138.
In molecular biology, competing endogenous RNAs regulate other RNA transcripts by competing for shared microRNAs (miRNAs). Models for ceRNA regulation describe how changes in the expression of one or multiple miRNA targets alter the number of unbound miRNAs and lead to observable changes in miRNA activity - i.e., the abundance of other miRNA targets. Models of ceRNA regulation differ greatly. Some describe the kinetics of target-miRNA-target interactions, where changes in the expression of one target species sequester one miRNA species and lead to changes in the dysregulation of the other target species. Others attempt to model more realistic cellular scenarios, where multiple RNA targets are affecting multiple miRNAs and where each target pair is co-regulated by multiple miRNA species. Some models focus on mRNA 3' UTRs as targets, and others consider long non-coding RNA targets as well. It's evident that our molecular-biochemical understanding of ceRNA regulation remains incomplete.
Epigenetic regulation of neurogenesis is the role that epigenetics plays in the regulation of neurogenesis.














