
Sterol regulatory element-binding proteins (SREBPs) are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors. Unactivated SREBPs are attached to the nuclear envelope and endoplasmic reticulum membranes. In cells with low levels of sterols, SREBPs are cleaved to a water-soluble N-terminal domain that is translocated to the nucleus. These activated SREBPs then bind to specific sterol regulatory element DNA sequences, thus upregulating the synthesis of enzymes involved in sterol biosynthesis. Sterols in turn inhibit the cleavage of SREBPs and therefore synthesis of additional sterols is reduced through a negative feed back loop.

The miR-1 microRNA precursor is a small micro RNA that regulates its target protein's expression in the cell. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give products at ~22 nucleotides. In this case the mature sequence comes from the 3' arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA. In humans there are two distinct microRNAs that share an identical mature sequence, and these are called miR-1-1 and miR-1-2.
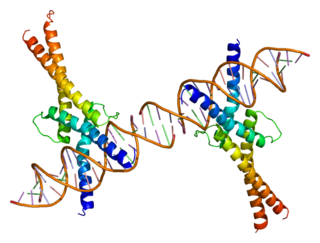
Sterol regulatory element-binding transcription factor 1 (SREBF1) also known as sterol regulatory element-binding protein 1 (SREBP-1) is a protein that in humans is encoded by the SREBF1 gene.

microRNA 21 also known as hsa-mir-21 or miRNA21 is a mammalian microRNA that is encoded by the MIR21 gene.
mir-127 microRNA is a short non-coding RNA molecule with interesting overlapping gene structure. miR-127 functions to regulate the expression levels of genes involved in lung development, placental formation and apoptosis. Aberrant expression of miR-127 has been linked to different cancers.

In molecular biology, mir-145 microRNA is a short RNA molecule that in humans is encoded by the MIR145 gene. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology, the miR-200 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by binding and cleaving mRNAs or inhibiting translation. The miR-200 family contains miR-200a, miR-200b, miR-200c, miR-141, and miR-429. There is growing evidence to suggest that miR-200 microRNAs are involved in cancer metastasis.

In molecular biology MicroRNA-223 (miR-223) is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. miR-223 is a hematopoietic specific microRNA with crucial functions in myeloid lineage development. It plays an essential role in promoting granulocytic differentiation while also being associated with the suppression of erythrocytic differentiation. miR-223 is commonly repressed in hepatocellular carcinoma and leukemia. Higher expression levels of miRNA-223 are associated with extranodal marginal-zone lymphoma of mucosa-associated lymphoid tissue of the stomach and recurrent ovarian cancer. In some cancers the microRNA-223 down-regulation is correlated with higher tumor burden, disease aggressiveness, and poor prognostic factors. MicroRNA-223 is also associated with rheumatoid arthritis, sepsis, type 2 diabetes, and hepatic ischemia.

In molecular biology, mir-221 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

miR-138 is a family of microRNA precursors found in animals, including humans. MicroRNAs are typically transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The excised region or, mature product, of the miR-138 precursor is the microRNA mir-138.

miR-33 is a family of microRNA precursors, which are processed by the Dicer enzyme to give mature microRNAs. miR-33 is found in several animal species, including humans. In some species there is a single member of this family which gives the mature product mir-33. In humans there are two members of this family called mir-33a and mir-33b, which are located in intronic regions within two protein-coding genes for Sterol regulatory element-binding proteins respectively.
In recent years it has become apparent that the environment and underlying mechanisms affect gene expression and the genome outside of the central dogma of biology. It has been found that many epigenetic mechanisms are involved in the regulation and expression of genes such as DNA methylation and chromatin remodeling. These epigenetic mechanisms are believed to be a contributing factor to pathological diseases such as type 2 diabetes. An understanding of the epigenome of Diabetes patients may help to elucidate otherwise hidden causes of this disease.
In molecular biology mir-365 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-370 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms. This microRNA, mir-370-3p, has been shown to play a role in heart failure. The upregulation of mir-370-3p in the sinus node leads to downregulation of the pacemaker ion channel, HCN4, and thus downregulation of the corresponding ionic current, which causes sinus bradycardia.
In molecular biology mir-153 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-590 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-872 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-938 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.

MicroRNA 34a (miR-34a) is a MicroRNA that in humans is encoded by the MIR34A gene.












