An allele, or allelomorph, is a variant of the sequence of nucleotides at a particular location, or locus, on a DNA molecule.

In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and the second is called recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes (autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked; these have an inheritance and presentation pattern that depends on the sex of both the parent and the child. Since there is only one copy of the Y chromosome, Y-linked traits cannot be dominant or recessive. Additionally, there are other forms of dominance, such as incomplete dominance, in which a gene variant has a partial effect compared to when it is present on both chromosomes, and co-dominance, in which different variants on each chromosome both show their associated traits.
Population genetics is a subfield of genetics that deals with genetic differences within and among populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as adaptation, speciation, and population structure.
Allele frequency, or gene frequency, is the relative frequency of an allele at a particular locus in a population, expressed as a fraction or percentage. Specifically, it is the fraction of all chromosomes in the population that carry that allele over the total population or sample size. Microevolution is the change in allele frequencies that occurs over time within a population.

In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative phenotypes, in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population.
Disassortative mating is a mating pattern in which individuals with dissimilar phenotypes mate with one another more frequently than would be expected under random mating. Disassortative mating reduces the mean genetic similarities within the population and produces a greater number of heterozygotes. The pattern is character specific, but does not affect allele frequencies. This nonrandom mating pattern will result in deviation from the Hardy-Weinberg principle.

A haplotype is a group of alleles in an organism that are inherited together from a single parent.
A quantitative trait locus (QTL) is a locus that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers correlate with an observed trait. This is often an early step in identifying the actual genes that cause the trait variation.
Balancing selection refers to a number of selective processes by which multiple alleles are actively maintained in the gene pool of a population at frequencies larger than expected from genetic drift alone. Balancing selection is rare compared to purifying selection. It can occur by various mechanisms, in particular, when the heterozygotes for the alleles under consideration have a higher fitness than the homozygote. In this way genetic polymorphism is conserved.
In biochemistry, isozymes are enzymes that differ in amino acid sequence but catalyze the same chemical reaction. Isozymes usually have different kinetic parameters, or are regulated differently. They permit the fine-tuning of metabolism to meet the particular needs of a given tissue or developmental stage.
Genetic association is when one or more genotypes within a population co-occur with a phenotypic trait more often than would be expected by chance occurrence.

Avena sterilis is a species of grass weed whose seeds are edible. Many common names of this plant refer to the movement of its panicle in the wind.

In population genetics, an ancestry-informative marker (AIM) is a single-nucleotide polymorphism that exhibits substantially different frequencies between different populations. A set of many AIMs can be used to estimate the proportion of ancestry of an individual derived from each population.
Dr George B. Johnson is a science educator who for many years has written a weekly column "On Science" in the St. Louis Post-Dispatch. For over 30 years he was a biology professor at Washington University and a genetics professor at Washington University School of Medicine. He has authored 44 scientific papers and ten high school and college biology texts. Over 3 million students have learned biology from these texts.
In genetics, transgressive segregation is the formation of extreme phenotypes, or transgressive phenotypes, observed in segregated hybrid populations compared to phenotypes observed in the parental lines. The appearance of these transgressive (extreme) phenotypes can be either positive or negative in terms of fitness. If both parents' favorable alleles come together, it will result in a hybrid having a higher fitness than the two parents. The hybrid species will show more genetic variation and variation in gene expression than their parents. As a result, the hybrid species will have some traits that are transgressive (extreme) in nature. Transgressive segregation can allow a hybrid species to populate different environments/niches in which the parent species do not reside, or compete in the existing environment with the parental species.
Marker assisted selection or marker aided selection (MAS) is an indirect selection process where a trait of interest is selected based on a marker linked to a trait of interest, rather than on the trait itself. This process has been extensively researched and proposed for plant- and animal- breeding.

Taste receptor 2 member 38 is a protein that in humans is encoded by the TAS2R38 gene. TAS2R38 is a bitter taste receptor; varying genotypes of TAS2R38 influence the ability to taste both 6-n-propylthiouracil (PROP) and phenylthiocarbamide (PTC). Though it has often been proposed that varying taste receptor genotypes could influence tasting ability, TAS2R38 is one of the few taste receptors shown to have this function.
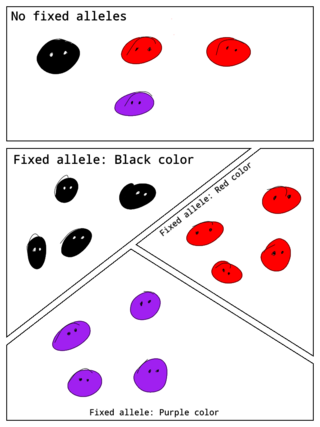
In population genetics, a fixed allele is an allele that is the only variant that exists for that gene in a population. A fixed allele is homozygous for all members of the population. The process by which alleles become fixed is called fixation.

Zygosity is the degree to which both copies of a chromosome or gene have the same genetic sequence. In other words, it is the degree of similarity of the alleles in an organism.

Robert Wayne Allard was an American plant breeder and plant population geneticist who is widely regarded as one of the leading plant population geneticists of the 20th century. Allard became chair of the genetics department at University of California, Davis in 1967; he was elected to the National Academy of Sciences in 1973, and was awarded the DeKalb-Pfizer Distinguished Career Award and the Crop Science Science of America Award. He was honored as the Nilsson-Ehle Lecturer of the Mendelian Society of Sweden and as the Wilhelmine Key lecturer of the American Genetic Association. He also served as president of the Genetics Society of America, the American Genetic Association and the American Society of Naturalists.










