Related Research Articles

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.
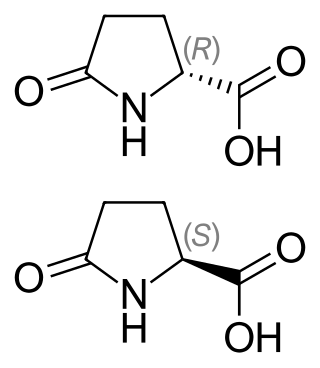
Pyroglutamic acid is a ubiquitous but understudied natural amino acid derivative in which the free amino group of glutamic acid or glutamine cyclizes to form a lactam. The names of pyroglutamic acid conjugate base, anion, salts, and esters are pyroglutamate, 5-oxoprolinate, or pidolate.
Nonribosomal peptides (NRP) are a class of peptide secondary metabolites, usually produced by microorganisms like bacteria and fungi. Nonribosomal peptides are also found in higher organisms, such as nudibranchs, but are thought to be made by bacteria inside these organisms. While there exist a wide range of peptides that are not synthesized by ribosomes, the term nonribosomal peptide typically refers to a very specific set of these as discussed in this article.

Membrane alanyl aminopeptidase also known as alanyl aminopeptidase (AAP) or aminopeptidase N (AP-N) is an enzyme that in humans is encoded by the ANPEP gene.

Cathepsin C (CTSC) also known as dipeptidyl peptidase I (DPP-I) is a lysosomal exo-cysteine protease belonging to the peptidase C1 protein family, a subgroup of the cysteine cathepsins. In humans, it is encoded by the CTSC gene.
Dipeptidyl peptidase-4 (DPP4), also known as adenosine deaminase complexing protein 2 or CD26 is a protein that, in humans, is encoded by the DPP4 gene. DPP4 is related to FAP, DPP8, and DPP9. The enzyme was discovered in 1966 by Hopsu-Havu and Glenner, and as a result of various studies on chemism, was called dipeptidyl peptidase IV [DP IV].
Aminopeptidases are enzymes that catalyze the cleavage of amino acids from the amino terminus (N-terminus) of proteins or peptides (exopeptidases). They are widely distributed throughout the animal and plant kingdoms and are found in many subcellular organelles, in cytosol, and as membrane components. Aminopeptidases are used in essential cellular functions. Many, but not all, of these peptidases are zinc metalloenzymes.

N-Acetylaspartylglutamic acid is a peptide neurotransmitter and the third-most-prevalent neurotransmitter in the mammalian nervous system. NAAG consists of N-acetylaspartic acid (NAA) and glutamic acid coupled via a peptide bond.

Leucyl aminopeptidases are enzymes that preferentially catalyze the hydrolysis of leucine residues at the N-terminus of peptides and proteins. Other N-terminal residues can also be cleaved, however. LAPs have been found across superkingdoms. Identified LAPs include human LAP, bovine lens LAP, porcine LAP, Escherichia coli LAP, and the solanaceous-specific acidic LAP (LAP-A) in tomato.

Dipeptidyl-peptidase 3 is an enzyme that in humans is encoded by the DPP3 gene.
Type 1 tumor necrosis factor receptor shedding aminopeptidase regulator, also known as endoplasmic reticulum aminopeptidase 1 (ARTS-1), is a protein which in humans is encoded by the ARTS-1 gene.

Puromycin-sensitive amino peptidase also known as cytosol alanyl aminopeptidase or alanine aminopeptidase (AAP) is an enzyme that in humans is encoded by the NPEPPS gene. It is used as a biomarker to detect damage to the kidneys, and that may be used to help diagnose certain kidney disorders. It is found at high levels in the urine when there are kidney problems.

Aspartyl aminopeptidase is an enzyme that in humans is encoded by the DNPEP gene.

The PGPEP1 gene in humans encodes the enzyme Pyroglutamyl-peptidase I.
Methionyl aminopeptidase is an enzyme. This enzyme catalyses the following chemical reaction
Pyroglutamyl-peptidase II is an enzyme. This enzyme catalyses the following chemical reaction

Scytalidocarboxyl peptidase B, also known as Scytalidoglutamic peptidase and Scytalidopepsin B is a proteolytic enzyme. It was previously thought to be an aspartic protease, but determination of its molecular structure showed it to belong a novel group of proteases, glutamic protease.

Glutamic proteases are a group of proteolytic enzymes containing a glutamic acid residue within the active site. This type of protease was first described in 2004 and became the sixth catalytic type of protease. Members of this group of protease had been previously assumed to be an aspartate protease, but structural determination showed it to belong to a novel protease family. The first structure of this group of protease was scytalidoglutamic peptidase, the active site of which contains a catalytic dyad, glutamic acid (E) and glutamine (Q), which give rise to the name eqolisin. This group of proteases are found primarily in pathogenic fungi affecting plant and human.
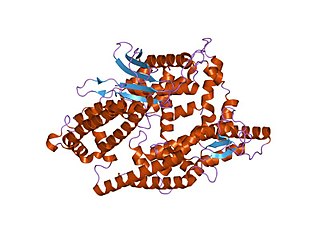
Thimet oligopeptidases, also known as TOPs, are a type of M3 metallopeptidases. These enzymes can be found in animals and plants, showing distinctive functions. In animals and humans, they are involved in the degradation of peptides, such as bradykinin, neurotensin, angiotensin I, and Aβ peptide, helping to regulate physiological processes. In plants, their role is related to the degradation of targeting peptides and the immune response to pathogens through Salicylic Acid (SA)-dependent stress signaling. In Arabidopsis thaliana—recognized as a model plant for scientific studies—two thimet oligopeptidases, known as TOP1 and TOP2, have been identified as targets for salicylic acid binding in the plant. These TOP enzymes are key components to understand the SA-mediated signaling where interactions exist with different components and most of the pathways are unknown.

The sedolisin family of peptidases are a family of serine proteases structurally related to the subtilisin (S8) family. Well-known members of this family include sedolisin ("pseudomonalisin") found in Pseudomonas bacteria, xanthomonalisin ("sedolisin-B"), physarolisin as well as animal tripeptidyl peptidase I. It is also known as sedolysin or serine-carboxyl peptidase. This group of enzymes contains a variation on the catalytic triad: unlike S8 which uses Ser-His-Asp, this group runs on Ser-Glu-Asp, with an additional acidic residue Asp in the oxyanion hole.
References
- ↑ Panosian KJ, Edberg SC (March 1989). "Measurement of constitutive L-pyrrolidonyl peptidase activity from Streptococcus and Enterococcus using tetrazotized 0-dianisidine". Antonie van Leeuwenhoek. 55 (3): 269–75. doi:10.1007/BF00393855. PMID 2569292.
- ↑ Tsuru D, Nakamura K, Yoshimoto T, Fujiwara K (1984). "Pyroglutamyl-peptidase from Bacillus amyloliquefaciens. An improved purification method and some properties of the enzyme". Biochim. Biophys. Acta. 791 (2): 117–122. doi:10.1016/0167-4838(84)90001-3.
- ↑ Awadé AC, Cleuziat P, Gonzalès T, Robert-Baudouy J (September 1994). "Pyrrolidone carboxyl peptidase (Pcp): an enzyme that removes pyroglutamic acid (pGlu) from pGlu-peptides and pGlu-proteins". Proteins. 20 (1): 34–51. doi:10.1002/prot.340200106. PMID 7824521.
- ↑ Patti JM, Schneider A, Garza N, Boles JO (December 1995). "Isolation and characterization of pcp, a gene encoding a pyrrolidone carboxyl peptidase in Staphylococcus aureus". Gene. 166 (1): 95–9. doi:10.1016/0378-1119(95)00561-0. PMID 8529900.
- ↑ Le Saux O, Gonzales T, Robert-Baudouy J (June 1996). "Mutational analysis of the active site of Pseudomonas fluorescens pyrrolidone carboxyl peptidase". Journal of Bacteriology. 178 (11): 3308–13. PMC 178084 . PMID 8655512.