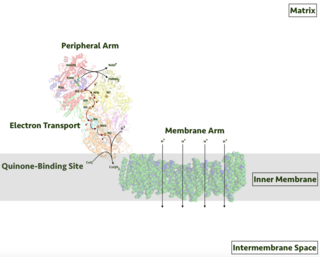
Respiratory complex I, EC 7.1.1.2 is the first large protein complex of the respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria.

The Rossmann fold is a tertiary fold found in proteins that bind nucleotides, such as enzyme cofactors FAD, NAD+, and NADP+. This fold is composed of alternating beta strands and alpha helical segments where the beta strands are hydrogen bonded to each other forming an extended beta sheet and the alpha helices surround both faces of the sheet to produce a three-layered sandwich. The classical Rossmann fold contains six beta strands whereas Rossmann-like folds, sometimes referred to as Rossmannoid folds, contain only five strands. The initial beta-alpha-beta (bab) fold is the most conserved segment of the Rossmann fold. The motif is named after Michael Rossmann who first noticed this structural motif in the enzyme lactate dehydrogenase in 1970 and who later observed that this was a frequently occurring motif in nucleotide binding proteins.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Flavoproteins are proteins that contain a nucleic acid derivative of riboflavin. These proteins are involved in a wide array of biological processes, including removal of radicals contributing to oxidative stress, photosynthesis, and DNA repair. The flavoproteins are some of the most-studied families of enzymes.

Thromboxane A synthase 1 , also known as TBXAS1, is a cytochrome P450 enzyme that, in humans, is encoded by the TBXAS1 gene.

Heme oxygenase, or haem oxygenase, is an enzyme that catalyzes the degradation of heme to produce biliverdin, ferrous ion, and carbon monoxide.
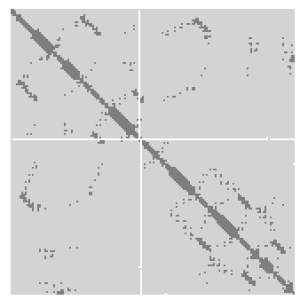
A protein contact map represents the distance between all possible amino acid residue pairs of a three-dimensional protein structure using a binary two-dimensional matrix. For two residues and , the element of the matrix is 1 if the two residues are closer than a predetermined threshold, and 0 otherwise. Various contact definitions have been proposed: The distance between the Cα-Cα atom with threshold 6-12 Å; distance between Cβ-Cβ atoms with threshold 6-12 Å ; and distance between the side-chain centers of mass.

Biliverdin reductase (BVR) is an enzyme found in all tissues under normal conditions, but especially in reticulo-macrophages of the liver and spleen. BVR facilitates the conversion of biliverdin to bilirubin via the reduction of a double bond between the second and third pyrrole ring into a single bond.

Lanosterol 14α-demethylase (CYP51A1) is the animal version of a cytochrome P450 enzyme that is involved in the conversion of lanosterol to 4,4-dimethylcholesta-8(9),14,24-trien-3β-ol. The cytochrome P450 isoenzymes are a conserved group of proteins that serve as key players in the metabolism of organic substances and the biosynthesis of important steroids, lipids, and vitamins in eukaryotes. As a member of this family, lanosterol 14α-demethylase is responsible for an essential step in the biosynthesis of sterols. In particular, this protein catalyzes the removal of the C-14α-methyl group from lanosterol. This demethylation step is regarded as the initial checkpoint in the transformation of lanosterol to other sterols that are widely used within the cell.

In enzymology, a shikimate dehydrogenase (EC 1.1.1.25) is an enzyme that catalyzes the chemical reaction
In enzymology, a cholestanetriol 26-monooxygenase (EC 1.14.13.15) is an enzyme that catalyzes the chemical reaction
In enzymology, a ferric-chelate reductase (EC 1.16.1.7) is an enzyme that catalyzes the chemical reaction
Flavin reductase a class of enzymes. There are a variety of flavin reductases, which bind free flavins and through hydrogen bonding, catalyze the reduction of these molecules to a reduced flavin. Riboflavin, or vitamin B, and flavin mononucleotide are two of the most well known flavins in the body and are used in a variety of processes which include metabolism of fat and ketones and the reduction of methemoglobin in erythrocytes. Flavin reductases are similar and often confused for ferric reductases because of their similar catalytic mechanism and structures.
In enzymology, a leghemoglobin reductase (EC 1.6.2.6) is an enzyme that catalyzes the chemical reaction

In enzymology, a NADPH—hemoprotein reductase is an enzyme that catalyzes the chemical reaction

Aldo-keto reductase family 1 member C3 (AKR1C3), also known as 17β-hydroxysteroid dehydrogenase type 5 is a key steroidogenic enzyme that in humans is encoded by the AKR1C3 gene.

Aldo-keto reductase family 1 member C1 also known as 20α-hydroxysteroid dehydrogenase, 3α-hydroxysteroid dehydrogenase, and dihydrodiol dehydrogenase 1/2 is an enzyme that in humans is encoded by the AKR1C1 gene.

3α-Hydroxysteroid dehydrogenase is an enzyme that in humans is encoded by the AKR1C4 gene. It is known to be necessary for the synthesis of the endogenous neurosteroids allopregnanolone, THDOC, and 3α-androstanediol. It is also known to catalyze the reversible conversion of 3α-androstanediol (5α-androstane-3α,17β-diol) to dihydrotestosterone and vice versa.
Nitric oxide reductase (NAD(P), nitrous oxide-forming) (EC 1.7.1.14, fungal nitric oxide reductase, cytochrome P450nor, NOR (ambiguous)) is an enzyme with systematic name nitrous oxide:NAD(P) oxidoreductase. This enzyme catalyses the following chemical reaction
Biliverdin reductase A is a protein that in humans is encoded by the BLVRA gene.

















