
In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Kynureninase or L-Kynurenine hydrolase (KYNU) is a PLP dependent enzyme that catalyses the cleavage of kynurenine (Kyn) into anthranilic acid (Ant). It can also act on 3-hydroxykynurenine and some other (3-arylcarbonyl)-alanines. Humans express one kynureninase enzyme that is encoded by the KYNU gene located on chromosome 2.

l-Kynurenine is a metabolite of the amino acid l-tryptophan used in the production of niacin.
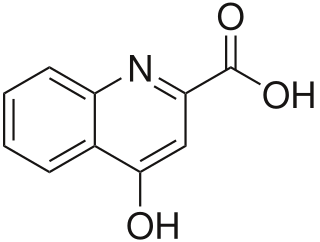
Kynurenic acid is a product of the normal metabolism of amino acid L-tryptophan. It has been shown that kynurenic acid possesses neuroactive activity. It acts as an antiexcitotoxic and anticonvulsant, most likely through acting as an antagonist at excitatory amino acid receptors. Because of this activity, it may influence important neurophysiological and neuropathological processes. As a result, kynurenic acid has been considered for use in therapy in certain neurobiological disorders. Conversely, increased levels of kynurenic acid have also been linked to certain pathological conditions.

An aromatic amino acid is an amino acid that includes an aromatic ring.

Squalene monooxygenase is a eukaryotic enzyme that uses NADPH and diatomic oxygen to oxidize squalene to 2,3-oxidosqualene. Squalene epoxidase catalyzes the first oxygenation step in sterol biosynthesis and is thought to be one of the rate-limiting enzymes in this pathway. In humans, squalene epoxidase is encoded by the SQLE gene. Several eukaryote genomes lack a squalene monooxygenase encoding gene, but instead encode an alternative squalene epoxidase that performs the same task.

The enzyme 4-hydroxybenzoate 3-monooxygenase, also commonly referred to as para-hydroxybenzoate hydroxylase (PHBH), is a flavoprotein belonging to the family of oxidoreductases. Specifically, it is a hydroxylase, and is one of the most studied enzymes and catalyzes reactions involved in soil detoxification, metabolism, and other biosynthetic processes.

4-hydroxyphenylacetate 3-monooxygenase (EC 1.14.14.9) is an enzyme that catalyzes the chemical reaction
In enzymology, a salicylate 1-monooxygenase (EC 1.14.13.1) is an enzyme that catalyzes the chemical reaction

Monooxygenases are enzymes that incorporate one hydroxyl group (−OH) into substrates in many metabolic pathways. In this reaction, the two atoms of dioxygen are reduced to one hydroxyl group and one H2O molecule by the concomitant oxidation of NAD(P)H. One important subset of the monooxygenases, the cytochrome P450 omega hydroxylases, is used by cells to metabolize arachidonic acid (i.e. eicosatetraenoic acid) to the cell signaling molecules, 20-hydroxyeicosatetraenoic acid or to reduce or totally inactivate the activate signaling molecules for example by hydroxylating leukotriene B4 to 20-hydroxy-leukotriene B5, 5-hydroxyeicosatetraenoic acid to 5,20-dihydroxyeicosatetraenoic acid, 5-oxo-eicosatetraenoic acid to 5-oxo-20-hydroxyeicosatetraenoic acid, 12-hydroxyeicosatetraenoic acid to 12,20-dihydroxyeicosatetraenoic acid, and epoxyeicosatrienoic acids to 20-hydroxy-epoxyeicosatrienoic acids.

Quinolinic acid, also known as pyridine-2,3-dicarboxylic acid, is a dicarboxylic acid with a pyridine backbone. It is a colorless solid. It is the biosynthetic precursor to niacin.

Tryptophan hydroxylase 1 (TPH1) is an isoenzyme of tryptophan hydroxylase which in humans is encoded by the TPH1 gene.

Kynurenine 3-monooxygenase is an enzyme that in humans is encoded by the KMO gene.

Hypertryptophanemia is a rare autosomal recessive metabolic disorder that results in a massive buildup of the amino acid tryptophan in the blood, with associated symptoms and tryptophanuria.
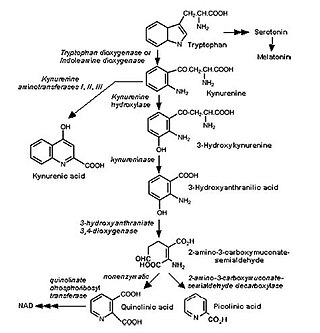
The kynurenine pathway is a metabolic pathway leading to the production of nicotinamide adenine dinucleotide (NAD+). Metabolites involved in the kynurenine pathway include tryptophan, kynurenine, kynurenic acid, xanthurenic acid, quinolinic acid, and 3-hydroxykynurenine. The kynurenine pathway is responsible for total catabolization of tryptophan about 95%. Disruption in the pathway is associated with certain genetic and psychiatric disorders.

Biopterin-dependent aromatic amino acid hydroxylases (AAAH) are a family of aromatic amino acid hydroxylase enzymes which includes phenylalanine 4-hydroxylase, tyrosine 3-hydroxylase, and tryptophan 5-hydroxylase. These enzymes primarily hydroxylate the amino acids L-phenylalanine, L-tyrosine, and L-tryptophan, respectively.
Tryptophan N-monooxygenase (EC 1.14.13.125, tryptophan N-hydroxylase, CYP79B1, CYP79B2, CYP79B3) is an enzyme with systematic name L-tryptophan,NADPH:oxygen oxidoreductase (N-hydroxylating). This enzyme catalyses the following chemical reaction
Tryptophan 7-halogenase (EC 1.14.19.9, PrnA, RebH) is an enzyme with systematic name L-tryptophan:FADH2 oxidoreductase (7-halogenating). This enzyme catalyses the following chemical reaction:
Cytochrome P450 omega hydroxylases, also termed cytochrome P450 ω-hydroxylases, CYP450 omega hydroxylases, CYP450 ω-hydroxylases, CYP omega hydroxylase, CYP ω-hydroxylases, fatty acid omega hydroxylases, cytochrome P450 monooxygenases, and fatty acid monooxygenases, are a set of cytochrome P450-containing enzymes that catalyze the addition of a hydroxyl residue to a fatty acid substrate. The CYP omega hydroxylases are often referred to as monoxygenases; however, the monooxygenases are CYP450 enzymes that add a hydroxyl group to a wide range of xenobiotic and naturally occurring endobiotic substrates, most of which are not fatty acids. The CYP450 omega hydroxylases are accordingly better viewed as a subset of monooxygenases that have the ability to hydroxylate fatty acids. While once regarded as functioning mainly in the catabolism of dietary fatty acids, the omega oxygenases are now considered critical in the production or break-down of fatty acid-derived mediators which are made by cells and act within their cells of origin as autocrine signaling agents or on nearby cells as paracrine signaling agents to regulate various functions such as blood pressure control and inflammation.

L-ornithine N5 monooxygenase (EC 1.14.13.195 or EC 1.14.13.196) is an enzyme which catalyzes one of the following chemical reactions:
L-ornithine + NADPH + O2 N(5)-hydroxy-L-ornithine + NADP+ + H2O L-ornithine + NAD(P)H + O2 N(5)-hydroxy-L-ornithine + NAD(P)+ + H2O


















