In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules.
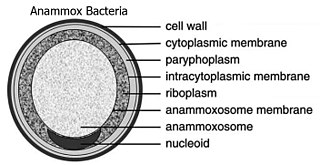
Planctomycetes are a phylum of aquatic bacteria and are found in samples of brackish, and marine and fresh water. They reproduce by budding. In structure, the organisms of this group are ovoid and have a holdfast, at the tip of a thin cylindrical extension from the cell body called the stalk, at the nonreproductive end that helps them to attach to each other during budding.

The Rickettsiales, informally called rickettsias, are an order of small Alphaproteobacteria. Some are notable pathogens, including Rickettsia, which causes a variety of diseases in humans, and Ehrlichia, which causes diseases in livestock. Another genus of well-known Rickettsiales is the Wolbachia, which infect about two-thirds of all arthropods and nearly all filarial nematodes. Genetic studies support the endosymbiotic theory according to which mitochondria and related organelles developed from members of this group.

The last universal common ancestor or last universal cellular ancestor (LUCA), also called the last universal ancestor (LUA), is the most recent population of organisms from which all organisms now living on Earth have a common descent—the most recent common ancestor of all current life on Earth. A related concept is that of progenote. LUCA is not thought to be the first life on Earth, but rather the only type of organism of its time to still have living descendants.

Alphaproteobacteria is a class of bacteria in the phylum Proteobacteria. Its members are highly diverse and possess few commonalities, but nevertheless share a common ancestor. Like all Proteobacteria, its members are gram-negative and some of its intracellular parasitic members lack peptidoglycan and are consequently gram variable.

Cobalamin riboswitch is a cis-regulatory element which is widely distributed in 5' untranslated regions of vitamin B12 (Cobalamin) related genes in bacteria. Riboswitches are metabolite binding domains within certain messenger RNAs (mRNAs) that serve as precision sensors for their corresponding targets. Allosteric rearrangement of mRNA structure is mediated by ligand binding, and this results in modulation of gene expression or translation of mRNA to yield a protein. Cobalamin in the form of adenosylcobalamin (Ado-CBL) is known to repress expression of proteins for vitamin B12 biosynthesis via a post-transcriptional regulatory mechanism that involves direct binding of Ado-CBL to 5' UTRs in relevant genes, preventing ribosome binding and translation of those genes. Before proof of riboswitch function, a conserved sequence motif called the B12 box was identified that corresponds to a part of the cobalamin riboswitch, and a more complete conserved structure was identified. Variants of the riboswitch consensus have been identified.
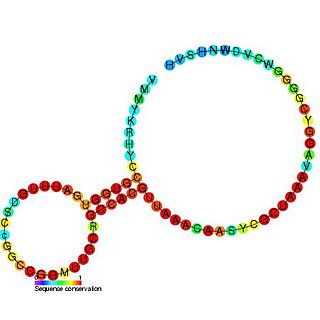
The SAM-II riboswitch is a RNA element found predominantly in alpha-proteobacteria that binds S-adenosyl methionine (SAM). Its structure and sequence appear to be unrelated to the SAM riboswitch found in Gram-positive bacteria. This SAM riboswitch is located upstream of the metA and metC genes in Agrobacterium tumefaciens, and other methionine and SAM biosynthesis genes in other alpha-proteobacteria. Like the other SAM riboswitch, it probably functions to turn off expression of these genes in response to elevated SAM levels. A significant variant of SAM-II riboswitches was found in Pelagibacter ubique and related marine bacteria and called SAM-V. Also, like many structured RNAs, SAM-II riboswitches can tolerate long loops between their stems.

Archaeal Richmond Mine acidophilic nanoorganisms (ARMAN) were first discovered in an extremely acidic mine located in northern California by Brett Baker in Jill Banfield's laboratory at the University of California Berkeley. These novel groups of archaea named ARMAN-1, ARMAN-2, and ARMAN-3 were missed by previous PCR-based surveys of the mine community because the ARMANs have several mismatches with commonly used PCR primers for 16S rRNA genes. Baker et al. detected them in a later study using shotgun sequencing of the community. The three groups were originally thought to represent three unique lineages deeply branched within the Euryarchaeota, a subgroup of the Archaea. However, based on a more complete archaeal genomic tree, they were assigned to a new superphylum named DPANN. The ARMAN groups now comprise deeply divergent phyla named Micrarchaeota and Parvarchaeota. Their 16S rRNA genes differ by as much as 17% between the three groups. Prior to their discovery, all of the Archaea shown to be associated with Iron Mountain belonged to the order Thermoplasmatales.
Mycoplasma laboratorium or Synthia refers to a synthetic strain of bacterium. The project to build the new bacterium has evolved since its inception. Initially the goal was to identify a minimal set of genes that are required to sustain life from the genome of Mycoplasma genitalium, and rebuild these genes synthetically to create a "new" organism. Mycoplasma genitalium was originally chosen as the basis for this project because at the time it had the smallest number of genes of all organisms analyzed. Later, the focus switched to Mycoplasma mycoides and took a more trial-and-error approach.

Archaea constitute a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria, but this classification is obsolete.
A wide variety of non-coding RNAs have been identified in various species of organisms known to science. However, RNAs have also been identified in "metagenomics" sequences derived from samples of DNA or RNA extracted from the environment, which contain unknown species. Initial work in this area detected homologs of known bacterial RNAs in such metagenome samples. Many of these RNA sequences were distinct from sequences within cultivated bacteria, and provide the potential for additional information on the RNA classes to which they belong.
The Pelagibacterales are an order in the Alphaproteobacteria composed of free-living marine bacteria that make up roughly one in three cells at the ocean's surface. Overall, members of the Pelagibacterales are estimated to make up between a quarter and a half of all prokaryotic cells in the ocean.

The crcB RNA motif is a conserved RNA structure identified by bioinformatics in a wide variety of bacteria and archaea. These RNAs were later shown to function as riboswitches that sense fluoride ions. These "fluoride riboswitches" increase expression of downstream genes when fluoride levels are elevated, and the genes are proposed to help mitigate the toxic effects of very high levels of fluoride.

The glutamine riboswitch is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria. It is also found in DNA extracted from uncultivated bacteria living in the ocean that are presumably species of cyanobacteria.
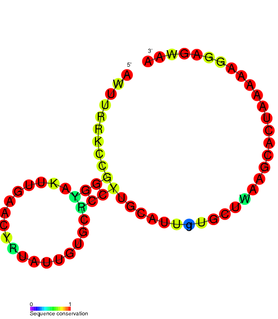
SAM-V riboswitch is the fifth known riboswitch to bind S-adenosyl methionine (SAM). It was first discovered in the marine bacterium Candidatus Pelagibacter ubique and can also be found in marine metagenomes. SAM-V features a similar consensus sequence and secondary structure as the binding site of SAM-II riboswitch, but bioinformatics scans cluster the two aptamers independently. These similar binding pockets suggest that the two riboswitches have undergone convergent evolution.

Marine microorganisms are defined by their habitat as microorganisms living in a marine environment, that is, in the saltwater of a sea or ocean or the brackish water of a coastal estuary. A microorganism is any microscopic living organism or virus, that is too small to see with the unaided human eye without magnification. Microorganisms are very diverse. They can be single-celled or multicellular and include bacteria, archaea, viruses and most protozoa, as well as some fungi, algae, and animals, such as rotifers and copepods. Many macroscopic animals and plants have microscopic juvenile stages. Some microbiologists also classify biologically active entities such as viruses and viroids as microorganisms, but others consider these as non-living.
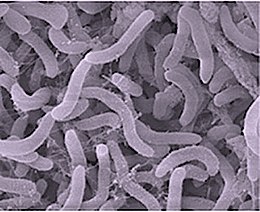
"Candidatus Scalindua" is a bacterial genus, and a proposed member of the order Planctomycetes. These bacteria lack peptidoglycan in their cell wall and have a compartmentalized cytoplasm. They are ammonium oxidizing bacteria found in marine environments.
HTVC010P is a virus which was discovered by Stephen Giovannoni and colleagues at Oregon State University. The Economist reports that a February 2013 paper in Nature says that "it probably really is the commonest organism on the planet". It is a bacteriophage that infects the extremely abundant bacteria Pelagibacter ubique in the Pelagibacterales order.
Genomic streamlining is a theory in evolutionary biology and microbial ecology that suggests that there is a reproductive benefit to prokaryotes having a smaller genome size with less non-coding DNA and fewer non-essential genes. There is a lot of variation in prokaryotic genome size, with the smallest free-living cell's genome being roughly ten times smaller than the largest prokaryote. Two of the bacterial taxa with the smallest genomes are Prochlorococcus and Pelagibacter ubique, both highly abundant marine bacteria commonly found in oligotrophic regions. Similar reduced genomes have been found in uncultured marine bacteria, suggesting that genomic streamlining is a common feature of bacterioplankton. This theory is typically used with reference to free-living organisms in oligotrophic environments.

Marine prokaryotes are marine bacteria and marine archaea. They are defined by their habitat as prokaryotes that live in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. All cellular life forms can be divided into prokaryotes and eukaryotes. Eukaryotes are organisms whose cells have a nucleus enclosed within membranes, whereas prokaryotes are the organisms that do not have a nucleus enclosed within a membrane. The three-domain system of classifying life adds another division: the prokaryotes are divided into two domains of life, the microscopic bacteria and the microscopic archaea, while everything else, the eukaryotes, become the third domain.