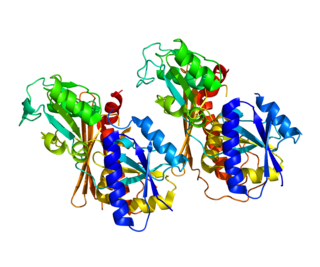
It has been suggested that this article be merged into Delta-aminolevulinic acid dehydratase . (Discuss) Proposed since February 2019. |
| porphobilinogen synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 DALA dehydratase | |||||||||
| Identifiers | |||||||||
| EC number | 4.2.1.24 | ||||||||
| CAS number | 9036-37-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Delta-aminolevulinic acid dehydratase | |
|---|---|
| Identifiers | |
| Symbol | ALAD |
| NCBI gene | 210 |
| HGNC | 395 |
| OMIM | 125270 |
| RefSeq | NM_001003945 |
| UniProt | P13716 |
| Other data | |
| EC number | 4.2.1.24 |
| Locus | Chr. 9 q32 |
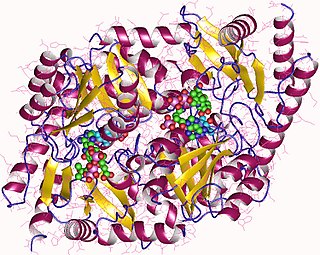
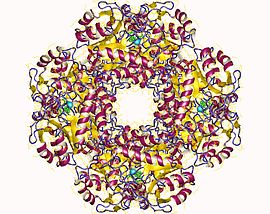
| ALAD | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 high resolution crystal structure of a mg2-dependent 5-aminolevulinic acid dehydratase | |||||||||
| Identifiers | |||||||||
| Symbol | ALAD | ||||||||
| Pfam | PF00490 | ||||||||
| Pfam clan | CL0036 | ||||||||
| InterPro | IPR001731 | ||||||||
| PROSITE | PDOC00153 | ||||||||
| SCOPe | 1aw5 / SUPFAM | ||||||||
| |||||||||
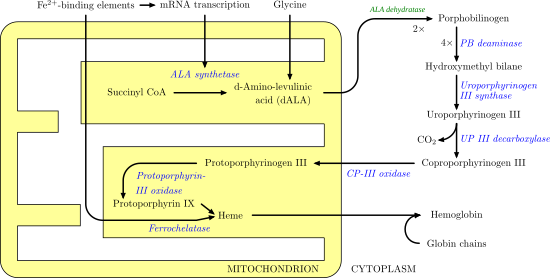
Porphobilinogen synthase (or ALA dehydratase, or aminolevulinate dehydratase) synthesizes porphobilinogen through the asymmetric condensation of two molecules of aminolevulinic acid. All natural tetrapyrroles, including hemes, chlorophylls and vitamin B12, share porphobilinogen as a common precursor.
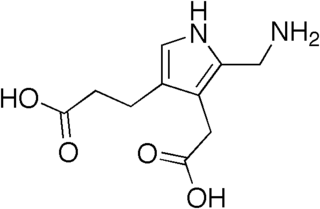
Porphobilinogen is an organic compound that occurs in living organisms as an intermediate in the biosynthesis of porphyrins, which include critical substances like hemoglobin and chlorophyll. The name is often abbreviated PBG.

Condensation is the change of the physical state of matter from the gas phase into the liquid phase, and is the reverse of vaporization. The word most often refers to the water cycle. It can also be defined as the change in the state of water vapor to liquid water when in contact with a liquid or solid surface or cloud condensation nuclei within the atmosphere. When the transition happens from the gaseous phase into the solid phase directly, the change is called deposition.
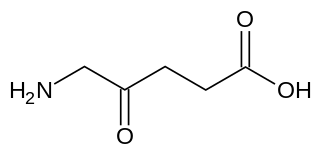
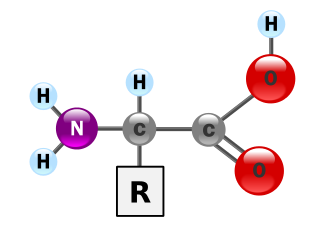
δ-Aminolevulinic acid, an endogenous non-proteinogenic amino acid, is the first compound in the porphyrin synthesis pathway, the pathway that leads to heme in mammals and chlorophyll in plants.
Contents
- Structure
- Allosteric regulators
- Intrinsic allosteric regulators
- Extrinsic allosteric regulators
- Deficiency
- PBGS as the prototype morpheein
- References
- External links
It catalyzes the second step of the biosynthesis of porphyrin:
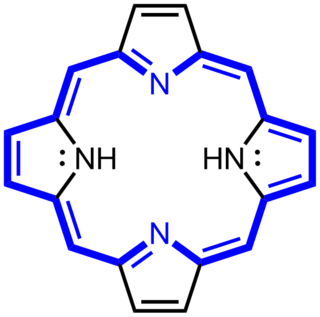
Porphyrins are a group of heterocyclic macrocycle organic compounds, composed of four modified pyrrole subunits interconnected at their α carbon atoms via methine bridges (=CH−). The parent porphyrin is porphine, a rare chemical compound of exclusively theoretical interest. Substituted porphines are called porphyrins. With a total of 26 π-electrons, of which 18 π-electrons form a planar, continuous cycle, the porphyrin ring structure is often described as aromatic. One result of the large conjugated system is that porphyrins typically absorb strongly in the visible region of the electromagnetic spectrum, i.e. they are deeply colored. The name "porphyrin" derives from the Greek word πορφύρα (porphyra), meaning purple.
- 2 δ-aminolevulinic acid porphobilinogen + 2 H2O
The porphobilinogen synthase catalyzed reaction is the first common step in the biosynthesis of all biological tetrapyrroles.
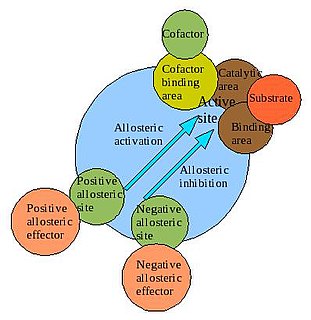
Porphobilinogen synthase is the prototype morpheein. [1]

Morpheeins are proteins that can form two or more different homo-oligomers, but must come apart and change shape to convert between forms. The alternate shape may reassemble to a different oligomer. The shape of the subunit dictates which oligomer is formed. Each oligomer has a finite number of subunits (stoichiometry). Morpheeins can interconvert between forms under physiological conditions and can exist as an equilibrium of different oligomers. These oligomers are physiologically relevant and are not misfolded protein; this distinguishes morpheeins from prions and amyloid. The different oligomers have distinct functionality. Interconversion of morpheein forms can be a structural basis for allosteric regulation. A mutation that shifts the normal equilibrium of morpheein forms can serve as the basis for a conformational disease. Features of morpheeins can be exploited for drug discovery. The dice image represents a morpheein equilibrium containing two different monomeric shapes that dictate assembly to a tetramer or a pentamer. The one protein that is established to function as a morpheein is porphobilinogen synthase, though there are suggestions throughout the literature that other proteins may function as morpheeins.