Related Research Articles

In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.
Mutagenesis is a process by which the genetic information of an organism is changed, resulting in a mutation. It may occur spontaneously in nature, or as a result of exposure to mutagens. It can also be achieved experimentally using laboratory procedures. A mutagen is a mutation-causing agent, be it chemical or physical, which results in an increased rate of mutations in an organism's genetic code. In nature mutagenesis can lead to cancer and various heritable diseases, but it is also a driving force of evolution. Mutagenesis as a science was developed based on work done by Hermann Muller, Charlotte Auerbach and J. M. Robson in the first half of the 20th century.
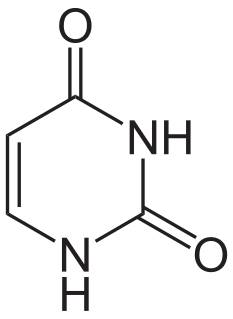
Uracil is one of the four nucleobases in the nucleic acid RNA that are represented by the letters A, G, C and U. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine. Uracil is a demethylated form of thymine.

Escherichia coli, also known as E. coli, is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms (endotherms). Most E. coli strains are harmless, but some serotypes (EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. The harmless strains are part of the normal microbiota of the gut, and can benefit their hosts by producing vitamin K2, (which helps blood to clot) and preventing colonisation of the intestine with pathogenic bacteria, having a symbiotic relationship. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for 3 days, but its numbers decline slowly afterwards.

The origin of replication is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semiconservative replication prior to cell division to ensure each daughter cell receives the full complement of chromosomes. This can either involve the replication of DNA in living organisms such as prokaryotes and eukaryotes, or that of DNA or RNA in viruses, such as double-stranded RNA viruses. Synthesis of daughter strands starts at discrete sites, termed replication origins, and proceeds in a bidirectional manner until all genomic DNA is replicated. Despite the fundamental nature of these events, organisms have evolved surprisingly divergent strategies that control replication onset. Although the specific replication origin organization structure and recognition varies from species to species, some common characteristics are shared.

The nucleoid is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a prokaryote is circular, and its length is very large compared to the cell dimensions needing it to be compacted in order to fit. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. Instead, the nucleoid forms by condensation and functional arrangement with the help of chromosomal architectural proteins and RNA molecules as well as DNA supercoiling. The length of a genome widely varies and a cell may contain multiple copies of it.

In biochemistry, suicide inhibition, also known as suicide inactivation or mechanism-based inhibition, is an irreversible form of enzyme inhibition that occurs when an enzyme binds a substrate analog and forms an irreversible complex with it through a covalent bond during the normal catalysis reaction. The inhibitor binds to the active site where it is modified by the enzyme to produce a reactive group that reacts irreversibly to form a stable inhibitor-enzyme complex. This usually uses a prosthetic group or a coenzyme, forming electrophilic alpha and beta unsaturated carbonyl compounds and imines.
A nick is a discontinuity in a double stranded DNA molecule where there is no phosphodiester bond between adjacent nucleotides of one strand typically through damage or enzyme action. Nicks allow DNA strands to untwist during replication, and are also thought to play a role in the DNA mismatch repair mechanisms that fix errors on both the leading and lagging daughter strands.
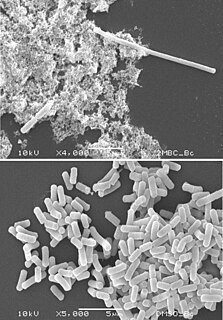
Filamentation is the anomalous growth of certain bacteria, such as Escherichia coli, in which cells continue to elongate but do not divide. The cells that result from elongation without division have multiple chromosomal copies. In the absence of antibiotics or other stressors, filamentation occurs at a low frequency in bacterial populations, the increased cell length protecting bacteria from protozoan predation and neutrophil phagocytosis by making ingestion of the cells more difficult. Filamentation is also a virulence factor thought to protect bacteria from antibiotics, and is associated with other aspects of bacterial virulence such as biofilm formation. The number and length of filaments within a bacterial population increases when the bacteria are treated with various chemical and physical agents. Some of the key genes involved in filamentation in E. coli include sulA and minCD.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.
Adaptive mutation, also called directed mutation or directed mutagenesis is a controversial evolutionary theory. It posits that mutations, or genetic changes, are much less random and more purposeful than traditional evolution, implying that can respond to environmental stresses by orthogenetically directing mutations to certain genes or areas of the genome. There have been a wide variety of experiments trying to prove the idea of adaptive mutation, at least in microorganisms.

In genetics, crosslinking of DNA occurs when various exogenous or endogenous agents react with two nucleotides of DNA, forming a covalent linkage between them. This crosslink can occur within the same strand (intrastrand) or between opposite strands of double-stranded DNA (interstrand). These adducts interfere with cellular metabolism, such as DNA replication and transcription, triggering cell death. These crosslinks can, however, be repaired through excision or recombination pathways.
Prokaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.

(p)ppGpp, guanosine pentaphosphate or tetraphosphate is an alarmone which is involved in the stringent response in bacteria, causing the inhibition of RNA synthesis when there is a shortage of amino acids present. This causes translation to decrease and the amino acids present are therefore conserved. Furthermore, ppGpp causes the up-regulation of many other genes involved in stress response such as the genes for amino acid uptake and biosynthesis.

Thymidine phosphorylase is an enzyme that is encoded by the TYMP gene and catalyzes the reaction:

A circular chromosome is a chromosome in bacteria, archaea, mitochondria, and chloroplasts, in the form of a molecule of circular DNA, unlike the linear chromosome of most eukaryotes.

A toxin-antitoxin system is a set of two or more closely linked genes that together encode both a "toxin" protein and a corresponding "antitoxin". Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies. When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK).
DNA Polymerase V is a polymerase enzyme involved in DNA repair mechanisms in prokaryotic bacteria, such as Escherichia coli. It is composed of a UmuD' homodimer and a UmuC monomer, forming the UmuD'2C protein complex. It is part of the Y-family of DNA Polymerases, which are capable of performing DNA translesion synthesis (TLS). Translesion polymerases bypass DNA damage lesions during DNA replication - if a lesion is not repaired or bypassed the replication fork can stall and lead to cell death. However, Y polymerases have low sequence fidelity during replication. When the UmuC and UmuD' proteins were initially discovered in E. coli, they were thought to be agents that inhibit faithful DNA replication and caused DNA synthesis to have high mutation rates after exposure to UV-light. The polymerase function of Pol V was not discovered until the late 1990s when UmuC was successfully extracted, consequent experiments unequivocally proved UmuD'2C is a polymerase. This finding lead to the detection of many Pol V orthologs and the discovery of the Y-family of polymerases.

The universal stress protein (USP) domain is a superfamily of conserved genes which can be found in bacteria, archaea, fungi, protozoa and plants. Proteins containing the domain are induced by many environmental stressors such as nutrient starvation, drought, extreme temperatures, high salinity, and the presence of uncouplers, antibiotics and metals.
Abraham Eisenstark was an American professor of microbiology. He was a Guggenheim Fellow for the academic year 1958–1959.
References
- 1 2 3 Ahmad, S. I.; Kirk, S. H.; Eisenstark, A. (October 1998). "Thymine Metabolism and Thymineless Death in Prokaryotes and Eukaryotes". Annual Review of Microbiology. 52: 591–625. doi:10.1146/annurev.micro.52.1.591. PMID 9891809.
- ↑ Longley, D. B.; Harkin, D. P.; Johnston, P. G. (2003). "5-Fluorouracil: Mechanisms of action and clinical strategies". Nature Reviews Cancer. 3 (5): 330–338. doi:10.1038/nrc1074. PMID 12724731. S2CID 4357553.
- ↑ Friedman, M. A.; Sadée, W. (1978). "The fluoropyrimidines: Biochemical mechanisms and design of clinical trials". Cancer Chemotherapy and Pharmacology. 1 (2): 77–82. doi:10.1007/bf00254040. PMID 373913. S2CID 10958670.
- ↑ Barner, H. D.; Cohen, S. S. (1954). "The Induction of Thymine Synthesis by T2 Infection of a Thymine Requiring Mutant of Escherichia Coli". Journal of Bacteriology. 68 (1): 80–88. doi:10.1128/JB.68.1.80-88.1954. PMC 357338 . PMID 13183905.
- 1 2 50 years ago in cell biology - A virologist recalls his work on cell growth inhibition
- ↑ Cohen, S. S.; Flaks, J. G.; Barner, H. D.; Loeb, M. R.; Lichtenstein, J. (1958). "The Mode of Action of 5-Fluorouracil and Its Derivatives". Proceedings of the National Academy of Sciences of the United States of America. 44 (10): 1004–1012. doi:10.1073/pnas.44.10.1004. PMC 528686 . PMID 16590300.
- ↑ Cohen, S. S.; Barner, H. D. (1954). "Studies on Unbalanced Growth in Escherichia Coli". Proceedings of the National Academy of Sciences of the United States of America. 40 (10): 885–893. doi:10.1073/pnas.40.10.885. PMC 534191 . PMID 16589586.
- 1 2 Martín, C. M.; Guzmán, E. C. (2011). "DNA replication initiation as a key element in thymineless death". DNA Repair. 10 (1): 94–101. doi:10.1016/j.dnarep.2010.10.005. PMID 21074501.
- 1 2 Nakayama, K.; Kusano, K.; Irino, N.; Nakayama, H. (1994). "Thymine starvation-induced structural changes in Escherichia coli DNA. Detection by pulsed field gel electrophoresis and evidence for involvement of homologous recombination". Journal of Molecular Biology. 243 (4): 611–620. doi:10.1016/0022-2836(94)90036-1. PMID 7966286.
- ↑ Sangurdekar, D. P.; Hamann, B. L.; Smirnov, D.; Srienc, F.; Hanawalt, P. C.; Khodursky, A. B. (2010). "Thymineless death is associated with loss of essential genetic information from the replication origin". Molecular Microbiology. 75 (6): 1455–1467. doi: 10.1111/j.1365-2958.2010.07072.x . PMID 20132444.
- ↑ Kuong, K. J.; Kuzminov, A. (2010). "Stalled replication fork repair and misrepair during thymineless death in Escherichia coli". Genes to Cells. 15 (6): 619–634. doi:10.1111/j.1365-2443.2010.01405.x. PMC 3965187 . PMID 20465561.
- ↑ Kuong, K. J.; Kuzminov, A (2012). "Disintegration of nascent replication bubbles during thymine starvation triggers RecA- and RecBCD-dependent replication origin destruction". The Journal of Biological Chemistry. 287 (28): 23958–70. doi:10.1074/jbc.M112.359687. PMC 3390671 . PMID 22621921.
- ↑ Sat, B.; Reches, M.; Engelberg-Kulka, H. (2003). "The Escherichia coli mazEF Suicide Module Mediates Thymineless Death". Journal of Bacteriology. 185 (6): 1803–1807. doi:10.1128/jb.185.6.1803-1807.2003. PMC 150121 . PMID 12618443.