Flaviviridae is a family of enveloped positive-strand RNA viruses which mainly infect mammals and birds. They are primarily spread through arthropod vectors. The family gets its name from the yellow fever virus; flavus is Latin for "yellow", and yellow fever in turn was named because of its propensity to cause jaundice in victims. There are 89 species in the family divided among four genera. Diseases associated with the group include: hepatitis (hepaciviruses), hemorrhagic syndromes, fatal mucosal disease (pestiviruses), hemorrhagic fever, encephalitis, and the birth defect microcephaly (flaviviruses).

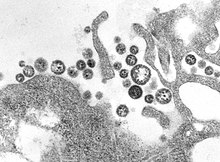
Rhabdoviridae is a family of negative-strand RNA viruses in the order Mononegavirales. Vertebrates, invertebrates, plants, fungi and protozoans serve as natural hosts. Diseases associated with member viruses include rabies encephalitis caused by the rabies virus, and flu-like symptoms in humans caused by vesiculoviruses. The name is derived from Ancient Greek rhabdos, meaning rod, referring to the shape of the viral particles. The family has 40 genera, most assigned to three subfamilies.
Rabies virus, scientific name Rabies lyssavirus, is a neurotropic virus that causes rabies in animals, including humans. Rabies transmission can occur through the saliva of animals and less commonly through contact with human saliva. Rabies lyssavirus, like many rhabdoviruses, has an extremely wide host range. In the wild it has been found infecting many mammalian species, while in the laboratory it has been found that birds can be infected, as well as cell cultures from mammals, birds, reptiles and insects. Rabies is reported in more than 150 countries and on all continents except Antarctica. The main burden of disease is reported in Asia and Africa, but some cases have been reported also in Europe in the past 10 years, especially in returning travellers.

Bunyavirales is an order of segmented negative-strand RNA viruses with mainly tripartite genomes. Member viruses infect arthropods, plants, protozoans, and vertebrates. It is the only order in the class Ellioviricetes. The name Bunyavirales derives from Bunyamwera, where the original type species Bunyamwera orthobunyavirus was first discovered. Ellioviricetes is named in honor of late virologist Richard M. Elliott for his early work on bunyaviruses.
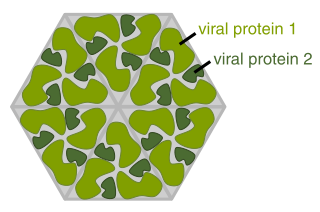
The term viral protein refers to both the products of the genome of a virus and any host proteins incorporated into the viral particle. Viral proteins are grouped according to their functions, and groups of viral proteins include structural proteins, nonstructural proteins, regulatory proteins, and accessory proteins. Viruses are non-living and do not have the means to reproduce on their own, instead depending on their host cell's machinery to do this. Thus, viruses do not code for most of the proteins required for their replication and the translation of their mRNA into viral proteins, but use proteins encoded by the host cell for this purpose.
Pseudoviridae is a family of viruses, which includes three genera.

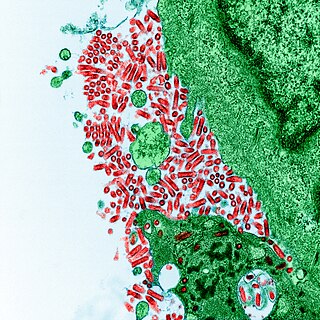
An arenavirus is a bi- or trisegmented ambisense RNA virus that is a member of the family Arenaviridae. These viruses infect rodents and occasionally humans. A class of novel, highly divergent arenaviruses, properly known as reptarenaviruses, have also been discovered which infect snakes to produce inclusion body disease. At least eight arenaviruses are known to cause human disease. The diseases derived from arenaviruses range in severity. Aseptic meningitis, a severe human disease that causes inflammation covering the brain and spinal cord, can arise from the lymphocytic choriomeningitis virus. Hemorrhagic fever syndromes, including Lassa fever, are derived from infections such as Guanarito virus, Junin virus, Lassa virus, Lujo virus, Machupo virus, Sabia virus, or Whitewater Arroyo virus. Because of the epidemiological association with rodents, some arenaviruses and bunyaviruses are designated as roboviruses.
Lymphocytic choriomeningitis (LCM) is a rodent-borne viral infectious disease that presents as aseptic meningitis, encephalitis or meningoencephalitis. Its causative agent is lymphocytic choriomeningitis mammarenavirus (LCMV), a member of the family Arenaviridae. The name was coined by Charles Armstrong in 1934.
Venezuelan hemorrhagic fever (VHF) is a zoonotic human illness first identified in 1989. The disease is most prevalent in several rural areas of central Venezuela and is caused by Guanarito mammarenavirus (GTOV) which belongs to the Arenaviridae family. The short-tailed cane mouse is the main host for GTOV which is spread mostly by inhalation of aerosolized droplets of saliva, respiratory secretions, urine, or blood from infected rodents. Person-to-person spread is possible, but uncommon.
The genome and proteins of HIV (human immunodeficiency virus) have been the subject of extensive research since the discovery of the virus in 1983. "In the search for the causative agent, it was initially believed that the virus was a form of the Human T-cell leukemia virus (HTLV), which was known at the time to affect the human immune system and cause certain leukemias. However, researchers at the Pasteur Institute in Paris isolated a previously unknown and genetically distinct retrovirus in patients with AIDS which was later named HIV." Each virion comprises a viral envelope and associated matrix enclosing a capsid, which itself encloses two copies of the single-stranded RNA genome and several enzymes. The discovery of the virus itself occurred two years following the report of the first major cases of AIDS-associated illnesses.

Argentinian mammarenavirus, better known as the Junin virus or Junín virus (JUNV), is an arenavirus in the Mammarenavirus genus that causes Argentine hemorrhagic fever (AHF). The virus took its original name from the city of Junín, around which the first cases of infection were reported, in 1958.

Orthobunyavirus is a genus of the Peribunyaviridae family in the order Bunyavirales. There are currently ~170 viruses recognised in this genus. These have been assembled into 103 species and 20 serogroups.
Plectrovirus is a genus of viruses, in the family Plectroviridae. Bacteria in the phylum Mycoplasmatota serve as natural hosts, making these viruses bacteriophages. Acholeplasma virus L51 is the only species in the genus.

Zaire ebolavirus, more commonly known as Ebola virus, is one of six known species within the genus Ebolavirus. Four of the six known ebolaviruses, including EBOV, cause a severe and often fatal hemorrhagic fever in humans and other mammals, known as Ebola virus disease (EVD). Ebola virus has caused the majority of human deaths from EVD, and was the cause of the 2013–2016 epidemic in western Africa, which resulted in at least 28,646 suspected cases and 11,323 confirmed deaths.
Batai orthobunyavirus (BATV) is a RNA virus belonging to order Bunyavirales, genus Orthobunyavirus.
Aquaparamyxovirus is a genus of viruses in the family Paramyxoviridae, order Mononegavirales. The genus includes two species. Fish serve as the natural hosts for AsaPV, in which the virus may cause proliferative gill inflammation.
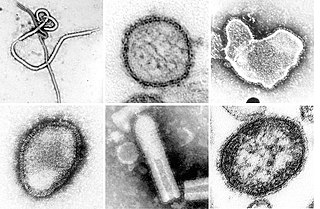
Negative-strand RNA viruses are a group of related viruses that have negative-sense, single-stranded genomes made of ribonucleic acid (RNA). They have genomes that act as complementary strands from which messenger RNA (mRNA) is synthesized by the viral enzyme RNA-dependent RNA polymerase (RdRp). During replication of the viral genome, RdRp synthesizes a positive-sense antigenome that it uses as a template to create genomic negative-sense RNA. Negative-strand RNA viruses also share a number of other characteristics: most contain a viral envelope that surrounds the capsid, which encases the viral genome, −ssRNA virus genomes are usually linear, and it is common for their genome to be segmented.
Bat mumps orthorubulavirus, formerly Bat mumps rubulavirus (BMV), is a member of genus Orthorubulavirus, family Paramyxoviridae, and order Mononegavirales. Paramyxoviridae viruses were first isolated from bats using heminested PCR with degenerate primers. This process was then followed by Sanger sequencing. A specific location of this virus is not known because it was isolated from bats worldwide. Although multiple paramyxoviridae viruses have been isolated worldwide, BMV specifically has not been isolated thus far. However, BMV was detected in African fruit bats, but no infectious form has been isolated to date. It is known that BMV is transmitted through saliva in the respiratory system of bats. While the virus was considered its own species for a few years, phylogenetic analysis has since shown that it is a member of Mumps orthorubulavirus.

Orthornavirae is a kingdom of viruses that have genomes made of ribonucleic acid (RNA), including genes which encode an RNA-dependent RNA polymerase (RdRp). The RdRp is used to transcribe the viral RNA genome into messenger RNA (mRNA) and to replicate the genome. Viruses in this kingdom share a number of characteristics which promote rapid evolution, including high rates of genetic mutation, recombination, and reassortment.
Rio Negro virus is an alphavirus that was first isolated in Argentina in 1980. The virus was first called Ag80-663 but was renamed to Rio Negro virus in 2005. It is a former member of the Venezuelan equine encephalitis complex (VEEC), which are a group of alphaviruses in the Americas that have the potential to emerge and cause disease. Río Negro virus was recently reclassified as a distinct species. Closely related viruses include Mucambo virus and Everglades virus.